













| General Information: |
|
| Name(s) found: |
vir-PA /
FBpp0071946
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1854 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][ISS]
integral to membrane [ISS] |
| Biological Process: |
RNA splicing, via transesterification reactions
[IMP]
primary sex determination, soma [NAS] dosage compensation [TAS] regulation of nuclear mRNA splicing, via spliceosome [TAS] border follicle cell migration [IMP] sex determination [TAS] |
| Molecular Function: |
nucleic acid binding
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MADVDDGSEL LFFDTFSHEE VTDINLDLVQ FPKPVFITQV RIIPLGARVQ ADFPGGVRLG 60
61 ATNPSKFDLE FFVNDLGMPA ASAFENLGLL RYNQNDCIHL DCSQEKIVTD GLVLRGWYST 120
121 ITLAIYGIFT NSVTEPIASP TLPCEPVGPE IANLSGEVLL QEDVLKDEWQ EPMQAELLTA 180
181 HKGNVSDYDP EDMEYGMSRD HYHQHAEEQE QREMRRLRRS THSTDHSPPP PRRSHTHSES 240
241 NDREYIRCSR DKGSRDWSRS PEYSSHRSRR KRSERSRSVV DEHKWPRTPP ASIDSPTRPR 300
301 SPDTMDYEDE DSRSHYKMQS SHYRHSSESL HRGERDRDDE DRSCTPQEQF EPILSDDEII 360
361 GDDEEDDAVD AAAIAEYERE LEAAAAAAPP AIDAFEPWQK PLLVFEGDMA AHFCKELETL 420
421 KLLFKKLVLQ TRCENVNAFS EEHGASVDER EQFVYLGEQL NNQLGYLAQH YKRRNFVLQQ 480
481 FFGNDELHLR QAANVLQIAL SFQAACMQPQ PAFKIRHIKL GARMAELLGS SEELFQHLLK 540
541 EHKFDIFEAV FRLYHEPYMA LSIKLQLLKA VYALLDTRMG IEHFMGAKNN GYQMIVEAIK 600
601 TAKLTRTKYA LQAIIKKLHL WEGLESVQIW CRRLFVDRII IPGNRDQMED TVITCQQIEF 660
661 AFEMLMDALF SSQLSYLQPR RFLPVSKKFE VVTDPTAQRS FGNALQSYLG QNSLAESLLV 720
721 MLANCKELPA TTYLSMLDLM HTLLRSHVGI DYFVDDAFPV TQTIVAILLG LDEVPRNPEE 780
781 KEEKAEKSDA EDKAMEVENE AVEAGGEKPT PPTADEEGKP VAAPISVPAP AAAPQVRPRP 840
841 ILRPVLPRLA RLGIEMSYKV QTRYHLDAIA YAAAAPEYDA VKLATHMHAI YSQTCDPAGR 900
901 QHTVEVLGLN NNLKIFMDLI KKEQRLQTQR QLSSPGTKYK SPVLSYAVDM VDACVRYCEQ 960
961 LDYLIEHGGV ILELAKNHET FEPSVSAVLQ EMYVYMKPLE AINVFVYDDI MPLVEVIGRS1020
1021 LDYLTTFPGD LIMAMRILRY LSISKPLAGQ KAPPVTEELK HRFVALQLYA ADGVQLCIQI1080
1081 MERLCAYFEQ PGAHAPALMT IQGVHCCQIM LPTLQILREL LSYAILCRDG TYKDLTAIDH1140
1141 LVKVYYLLYY FPTRCQAGPE VEQCKMEVVQ TLLAYTQPNE QDEESLHKSL WTLMIREVLK1200
1201 NVDGPAHFIP GLKLLAELLP LPLPMPQPLC DQLQQQHKQR LITERKLWSA HLHPQSGQIA1260
1261 KLVEALAPSS FPQLSELLQR VCMQLSDLAP NMTLLIAKTI TELLCNEYQT SNCIPTTNLE1320
1321 RLLRFSTRLC AFAPLKSSML SILSGKFWEL FQSLLALNEF NDVVSNCQEA VHRILDSFLD1380
1381 SGISLISHKS TASPALNLAA ALPPKELIPR IIDAVFSNLT SVEVTHGISI LAVRNLVILT1440
1441 EHDFTFYHLA QLLKQKITEF QAWMERVILH NETVEYNANI ESLILLLRSL TQIEPPPAMS1500
1501 AMPHRTLKLG ATELAQLVEF QDIELAKPPV LSRILTVMEK HKAVANEAAL SDLKQLILLQ1560
1561 ASKQEILAGT STETPPEAEG EANPSASSCS ASLTVEPYLP QAEGIVTQYE ARPIFTRFCA1620
1621 TAENAQLTAR YWLDPLPIEL IEDMNEPIYE RIACDLTDLA NVCLNPDLNV AGDSKRVMNL1680
1681 SGSPQSNREM TPTAPCFRTR RVEVEPATGR PEKKMFVSSV RGRGFARPPP SRGDLFRSRP1740
1741 PNTSRPPSLH VDDFLALETC GAQPTGPTGY NKIPSMLRGS RVGRNRGSRI SAAAAFRQKK1800
1801 MMRIGSPSSW AESPGSYRSA SDSHFSSSDS HYSSPHYSGR PRGRGLRSRP SYLR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
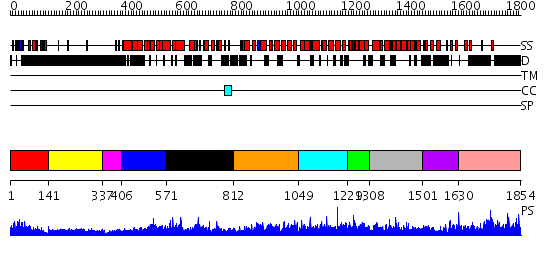
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..140] | 1.339969 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [141..336] | 7.415996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [337..405] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [406..570] | 8.363994 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [571..811] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [812..1048] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1049..1228] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1229..1307] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1308..1500] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1501..1629] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1630..1854] | 1.020972 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)