













| General Information: |
|
| Name(s) found: |
Tbp-PA /
FBpp0071596
[FlyBase]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 353 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[NAS]
transcription factor TFIID complex [IPI][ISS][NAS] transcription factor TFIIIB complex [IPI] DNA-directed RNA polymerase III complex [IC] |
| Biological Process: |
regulation of transcription, DNA-dependent
[IEA]
snRNA transcription from RNA polymerase III promoter [IDA] regulation of transcription from RNA polymerase III promoter [IMP] tRNA transcription from RNA polymerase III promoter [IDA][IMP] transcription initiation from RNA polymerase II promoter [ISS] regulation of transcription from RNA polymerase II promoter [NAS][TAS] |
| Molecular Function: |
DNA binding
[IEA][NAS]
protein binding [IPI] general RNA polymerase II transcription factor activity [ISS] transcription factor binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDQMLSPNFS IPSIGTPLHQ MEADQQIVAN PVYHPPAVSQ PDSLMPAPGS SSVQHQQQQQ 60
61 QSDASGGSGL FGHEPSLPLA HKQMQSYQPS ASYQQQQQQQ QLQSQAPGGG GSTPQSMMQP 120
121 QTPQSMMAHM MPMSERSVGG SGAGGGGDAL SNIHQTMGPS TPMTPATPGS ADPGIVPQLQ 180
181 NIVSTVNLCC KLDLKKIALH ARNAEYNPKR FAAVIMRIRE PRTTALIFSS GKMVCTGAKS 240
241 EDDSRLAARK YARIIQKLGF PAKFLDFKIQ NMVGSCDVKF PIRLEGLVLT HCNFSSYEPE 300
301 LFPGLIYRMV RPRIVLLIFV SGKVVLTGAK VRQEIYDAFD KIFPILKKFK KQS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
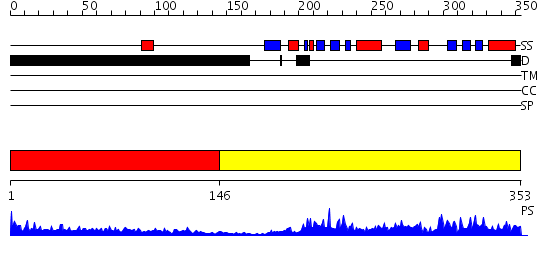
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..145] | 7.0 | Sec24 | |
| 2 | View Details | [146..353] | 87.09691 | Structure of a Yeast TFIIA/TBP/TATA-box DNA Complex |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)