













| General Information: |
|
| Name(s) found: |
gi|7302641
[NCBI NR]
gi|24654891 [NCBI NR] |
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 2280 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][NAS]
|
| Biological Process: |
negative regulation of transcription from RNA polymerase II promoter
[NAS]
regulation of imaginal disc growth [IMP] photoreceptor cell development [NAS] imaginal disc-derived wing morphogenesis [IMP] axon guidance [IMP][NAS] larval locomotory behavior [IMP][TAS] wing disc dorsal/ventral pattern formation [IGI] axon target recognition [IMP] negative regulation of gene-specific transcription from RNA polymerase II promoter [IMP] |
| Molecular Function: |
transcription factor activity
[NAS]
zinc ion binding [IEA] transcription corepressor activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEKKLAKVAL NSSNVATESN IKNNKLLANL SAAGAATATT ATTGTATTTT ATTANQALNF 60
61 NNKTKATANA TAASANNRHN NNNNSSAIKK HTNTKQLAGK SPACNSSSSS LSSSSSSNSS 120
121 ESKDTNFEYE DEWNIGGIPE LLDDLDADIE KSAHSSGGGN QATALNAKQA NSSSTSSSSS 180
181 SKGGASSSSS SAAATGSSSS HKSHKTTLHS NLSATSPTTI KFTRQPVAIG GANSSSSSSA 240
241 AAAPSGGSAN VVAKGSSSSS SSTSSSSSGK HHHHHHHHSN SSSSGSSSKG YKSALVAQLN 300
301 SPSPLNSNSK SLSGSGSGSG NTNGAAGAGA GSTLSSSTFA GFSKGGSLVS SSAGAAAALA 360
361 AGSGQQGSKF SAGGMSSQTG SGSGGNNTSN SNNSSSGSGG SGSGSSTGNT ASGSGNNNST 420
421 SAGGPPSSQG GNNGNGSGSS SSSSSGKSSA KMSIDHQATL DKGLKMKIKR TKPGTKSSEA 480
481 KHEIVKATDQ QQNGALGAGS NNSANEDGSS GSSSTNASSL GSTNSSSSAS SGSSSSSGSS 540
541 SSSSKKHLNN ASSGSGSSSS GGGSQNNASG HASGGGSSGS SQSTPQGTKR GSSGHRREKT 600
601 KDKNAHSNRM SVDKSAAAAS AAGEKDTPEK CSGTGAGGSP CSCNGDVGAP CSHHACIRRA 660
661 AHMSNSAGNA NSSAGTGQSG GSSSMSAVPP GVFTPSAGSP STGSPSTVVP AAASLLAATG 720
721 AASSSASQMA SSSAGGVGGS GGGANAPGPP GKESAGSIKI SSHIAAQLAA AAASNSYSGS 780
781 GANTNQGQNS NAGGNGGSES KAAAAAQAKL MAPGMISATM HHTISVPAGT GTGDDDTKSP 840
841 PAKRVKHEAG ASGAGGGKEM VDICIGTSVG TITEPDCLGP CEPGTSVTLE GIVWHETEGG 900
901 VLVVNVTWRG KTYVGTLLDC TRHDWAPPRF CDSPTEELDS RTPKGRGKRG RSAGLTPDLS 960
961 NFTETRSSIY FSHAQVHSKL RNGATKGRGG ATPSTSPTAF LPPRPEKRKS KDEAPSPLNG1020
1021 DASDGASVGG IGGAGGVNMV NASGIPISAS GGGLATQPQS LLNPVTGLNV QISTKKCKTA1080
1081 SPCAISPVLL ECPEQDCSKK YKHANGLRYH QSHAHGAGGG ASSMDEDSMQ APEDPATPPS1140
1141 PGVASGTGSG ASVASSAVPA TAPSAGQGTV AVSPNTPLAN SSNPVTNGNV APSAPATGSV1200
1201 TIAAPNTTPS VVETQAPLTG PPPVTPPAPT PICAVATPGA EQSVSSVLPL GNLPLTAGPN1260
1261 SATQQQQPPT QQQQPQLLVP GGSAASLQQQ QQQQQPVAGG SITAGISGQA LSQHQQQLMG1320
1321 GLPAMLSDQQ QQALLQQGAL KAGVLRFGPP DGNPLQQQPG QASVNPQTQQ SPPRPPSHVQ1380
1381 DQQTPSAYAQ QAGLKTSPGF GSVGVGAASS KQKKNRKSPG PSDFEGRVSR EDVQSPAYSD1440
1441 ISDDSTPVAE QEMLDKSVGQ AVTAKHIELM GKKPTEVGVG VPPPPAPNMY VPGMYQFYPA1500
1501 QQQSAPPPQQ QQQQPQYMVQ TEPGKPPGLP PALTQAQQQQ QLQPGAPPPT SQPPSHLLGP1560
1561 PGQQSVAAHL ADYSGKNKDP PLDLMTKPQP QPGQPPSQQQ QSGQLSGQEN NGKDVGPPTS1620
1621 QPGSQPPPVN LSAVAGPPPG SLPPGLGGLS ALGAAGLGGP GPGKGMPHFY PFNFIPPAYP1680
1681 YNVDPNFGSV SIVASEEAAK LSGHPGLPPS SQAQQLSGIS IKEERLKESP SPHDQPKHMP1740
1741 SQQQMIASKL IKQEPMTKQE IKQEPNSNPG QQHPPPQQQP APQPQQQQPP PPQPQQPHAL1800
1801 HPKDLQALGA YPAIYQRHSI NLAVQQAREE ELRRYYMFTG RQNSAAAAAA AAAQNAASGG1860
1861 LPPHPGMMHK DEPGMGSAAQ QQQQQQQQQM QIAQQQQQAI QQHHQHLQQQ HQAQQQQQQQ1920
1921 QHQQQQQQQQ QQQQQQQQQQ QQQKLKQSQA ASAGANNKAT NLTKDSPKQK GGDDDQPLKV1980
1981 KQEGQKPTME TQGPPPPPTS QYFLHPSYIS PTPFGFDPNH PMYRNVLMSA AGPYNTAPYH2040
2041 LPIPRPYHAP EDLSRNTGTK ALDALHHAAS QYYTTHKIHE LSERALKSPT SGSGPVKVSV2100
2101 SSPSIGPPQP GGPTSSGPGS GPVSGVLGPG SGSNQQPGSA PGSAGGVPLN LQPPPGGMGP2160
2161 SPGSKPDLSG PKGHGGVTPG SSLDGHKQSM PGGPPPNGPS GNGAVGGVGG AAGNGAAGGG2220
2221 GAGAADSRSP PPQRHVHTHH HTHVGLGYPM YPAPYGAAVL ASQQAAAVAV INPFPPGPSK2280
2281 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
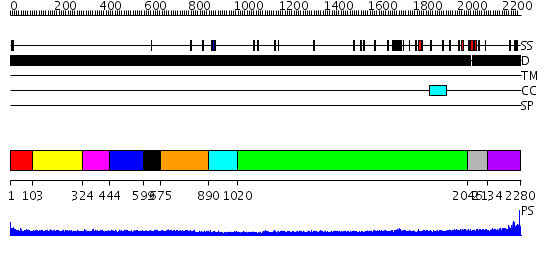
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..102] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [103..323] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [324..443] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [444..598] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [599..674] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [675..889] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [890..1019] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1020..2045] | 27.0 | No description for 1y0fB was found. | |
| 9 | View Details | [2046..2133] | 25.045757 | No description for 1y0fA was found. | |
| 10 | View Details | [2134..2280] | 2.34 | Crystal structure of the C-terminal domain of BclA, the major antigen of the exosporium of the Bacillus anthracis spore. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|