













| General Information: |
|
| Name(s) found: |
krimp-PA /
FBpp0086333
[FlyBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 746 amino acids |
Gene Ontology: |
|
| Cellular Component: |
perinuclear region of cytoplasm
[IDA]
P granule [IDA] |
| Biological Process: |
negative regulation of oskar mRNA translation
[IMP]
karyosome formation [IMP] oocyte dorsal/ventral axis specification [IMP] |
| Molecular Function: |
nucleic acid binding
[IEA]
zinc ion binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNLEDISMIM KLFDSNMHKL QGNLRSYQTE MHQIHKELTE KLSHADLLYR SLIPLHDHLV 60
61 ASLSEVNAHV MKLNVQLHIN RQSVRLGDYE YYEKSIDNPY SSIRSGLQAI EKPGCAEAIC 120
121 QSSKPAFVEC LPSSTSEEVP VVAVQEASST NQLDAISVVN ENLSEERDAT PQPLAVSKNM 180
181 EETMPSNPFH EQLEGSLEEI PVGSKIVVET EKANNPVRSE ASAPATSDNQ SLLAAKQGTQ 240
241 TIGTGICNKI SKSTINMPNN WQLENPTEVT AASIEKVNKL PKSPRNRFLL PPKGGTETTR 300
301 RDIYNQILKD MAAFPENTIV TAVLASVDVT DNCAYVAKWD ESSDRIKKVL QRQLPLQELD 360
361 QLPDYGDIFA VLDSINNIIT RITINSSSAG GGYDAYLIDF GEHIHFDGNE TIFKLPDDIK 420
421 RLPAQAIRCD LINCDIANMH CFVNTYIKIR VHENNNSTLV AEPVIDRLSR PTKTNTTKYP 480
481 AGITEDDMAM LNEIDESTSD PLKAVLGFRP KDEQRICRHY DPKLNGCFKG NNCRFAHEPF 540
541 APNGATKDVE LARALPETIF DTTVHFEIGS IVGILITFIN GPTEVYGQFL DGSPPLVWDK 600
601 KDVPENKRTF KSKPRLLDIV LALYSDGCFY RAQIIDEFPS EYMIFYVDYG NTEFVPLSCL 660
661 APCENVDSFK PHRVFSFHIE GIVRSKNLTH QKTIECIEYL KSKLLNTEMN VHLVQRLPDG 720
721 FLIRFLDDWK YIPEQLLQRN YAQVSQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
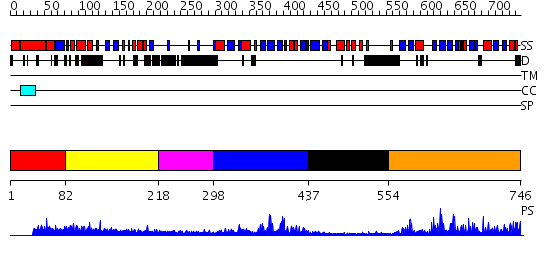
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..81] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [82..217] | 1.041995 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [218..297] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [298..436] | 10.39794 | No description for 2d9tA was found. | |
| 5 | View Details | [437..553] | 1.18 | No description for 2d9mA was found. | |
| 6 | View Details | [554..746] | 37.30103 | No description for 2hqeA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)