













| General Information: |
|
| Name(s) found: |
Amph-PA /
FBpp0086941
[FlyBase]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 602 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA]
synapse [IDA] plasma membrane [IDA] |
| Biological Process: |
synaptic vesicle endocytosis
[IEP][IMP][TAS]
rhabdomere development [IMP] regulation of muscle contraction [IMP] protein localization [IMP] rhabdomere membrane biogenesis [IMP] endocytosis [NAS] exocytosis [TAS] neurotransmitter secretion [NAS] |
| Molecular Function: |
protein binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTENKGIMLA KSVQKHAGRA KEKILQNLGK VDRTADEIFD DHLNNFNRQQ ASANRLQKEF 60
61 NNYIRCVRAA QAASKTLMDS VCEIYEPQWS GYDALQAQTG ASESLWADFA HKLGDQVLIP 120
121 LNTYTGQFPE MKKKVEKRNR KLIDYDGQRH SFQNLQANAN KRKDDVKLTK GREQLEEARR 180
181 TYEILNTELH DELPALYDSR ILFLVTNLQT LFATEQVFHN ETAKIYSELE AIVDKLATES 240
241 QRGSNTLRKQ TSNPIKTSSP VQSPVNKLNN ANINSNYQNQ ITTNGGSSLA NSPTSTSSSL 300
301 QEPRFDSVSS TPEPRPESPA AALVSATPSS PVENGVTTKS LERPELSGLN ASAKATTTTQ 360
361 TSPTEDKAVV SEAVKPSETE GAAVAASVTP APPATPAQIN GNNNEPSIVK EGGKQPKELP 420
421 STTSNAEAAA EAAANNGNSI EEHKQKKLGN DTTVTATETV TVTQHSVTST DTDNIVTISD 480
481 TNTDTDTKTS TGTSQKGRPV PVVNRHSVNN LNKNPFEDDD ERIYEVPADA NTADLPPGVL 540
541 YRVKATYGYA KEDVDELSFE IGDLIRVIEY DDPEDQEEGW LMGQKEGTNE KGLFPANFTR 600
601 PI |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
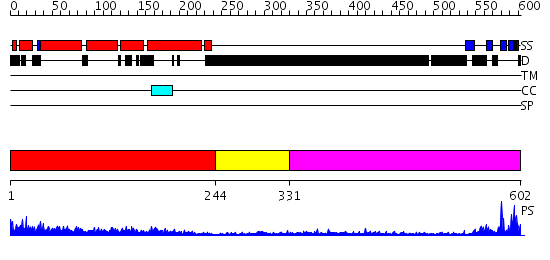
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..243] | 54.522879 | Amphiphysin BAR domain from Drosophila | |
| 2 | View Details | [244..330] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [331..602] | 15.30103 | NMR STRUCTURE OF THE TUMOR SUPPRESSOR BIN1: ALTERNATIVE SPLICING IN MELANOMA AND INTERACTION WITH C-MYC |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)