













| General Information: |
|
| Name(s) found: |
Su(var)2-10-PB /
FBpp0087658
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 593 amino acids |
Gene Ontology: |
|
| Cellular Component: |
polytene chromosome, telomeric region
[IDA]
nucleus [IDA][TAS] polytene chromosome [IDA] cytoplasm [IDA][TAS] nuclear lamina [IDA] nucleoplasm [IDA] |
| Biological Process: |
chromosome organization
[IMP][TAS]
hemopoiesis [TAS] chromosome condensation [IMP] regulation of protein catabolic process [TAS] regulation of JAK-STAT cascade [TAS] compound eye development [IMP][TAS] negative regulation of JAK-STAT cascade [NAS] imaginal disc growth [TAS] |
| Molecular Function: |
nucleic acid binding
[IEA]
DNA binding [ISS] zinc ion binding [IEA] DEAD/H-box RNA helicase binding [NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVQMLRVVEL QKILSFLNIS FAGRKTDLQS RILSFLRTNL ELLAPKVQEV YAQSVQEQNA 60
61 TLQYIDPTRM YSHIQLPPTV QPNPVGLVGS GQGVQVPGGQ MNVVGGAPFL HTHSINSQLP 120
121 IHPDVRLKKL AFYDVLGTLI KPSTLVPRNT QRVQEVPFYF TLTPQQATEI ASNRDIRNSS 180
181 KVEHAIQVQL RFCLVETSCD QEDCFPPNVN VKVNNKLCQL PNVIPTNRPN VEPKRPPRPV 240
241 NVTSNVKLSP TVTNTITVQW CPDYTRSYCL AVYLVKKLTS TQLLQRMKTK GVKPADYTRG 300
301 LIKEKLTEDA DCEIATTMLK VSLNCPLGKM KMLLPCRAST CSHLQCFDAS LYLQMNERKP 360
361 TWNCPVCDKP AIYDNLVIDG YFQEVLGSSL LKSDDTEIQL HQDGSWSTPG LRSETQILDT 420
421 PSKPAQKVEV ISDDIELISD DAKPVKRDLS PAQDEQPTST SNSETVDLTL SDSDDDMPLA 480
481 KRRPPAKQAV ASSTSNGSGG GQRAYTPAQQ PQQSESPEQQ ASRQSPEKQT VSEQQLQQQQ 540
541 HEQPATAAVH ASLLESLAAA VADQKHFQLL DLAAVAAAAA ATASSGQSQN AGP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
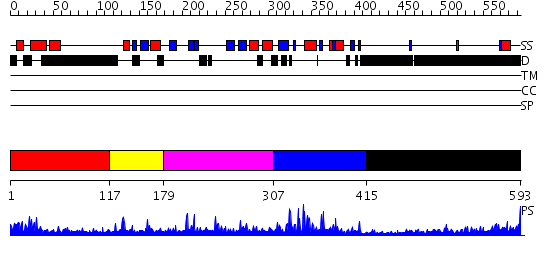
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..116] | 15.69897 | Solution structure of human p53 binding domain of PIAS-1 | |
| 2 | View Details | [117..178] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [179..306] | 1.125969 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [307..414] | 7.69897 | No description for 2yu4A was found. | |
| 5 | View Details | [415..593] | 9.140997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)