













| General Information: |
|
| Name(s) found: |
CSN4-PB /
FBpp0290448
[FlyBase]
CSN4-PA / FBpp0087935 [FlyBase] |
| Description(s) found:
Found 33 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 407 amino acids |
Gene Ontology: |
|
| Cellular Component: |
signalosome
[IMP][IPI][ISS][NAS][TAS]
|
| Biological Process: |
compound eye photoreceptor cell differentiation
[IMP]
protein deneddylation [IMP] protein stabilization [IMP] negative regulation of proteasomal ubiquitin-dependent protein catabolic process [IMP] |
| Molecular Function: |
protein binding
[IPI]
NEDD8 activating enzyme activity [IMP] promoter binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAANYGISTA ALRSQLMGLI NFTGTHKDQA DKYRQLLKTV LTNTGQELID GLRLFVEAIV 60
61 NEHVSLVISR QILNDVGSEL SKLPDDLSKM LSHFTLEKVN PRVISFEEQV AGIRFHLANI 120
121 YERNQQWRDA ATVLVGIPLE TGQKQYSVEC KLGTYLKIAR LYLEDNDSVQ AELFINRASL 180
181 LQAETNSEEL QVLYKVCYAR VLDYRRKFIE AAQRYNELSY RKIVDQGERM TALKKALICT 240
241 VLASAGQQRS RMLATLFKDE RCQHLPAYGI LEKMYLERII RRSELQEFEA LLQDHQKAAT 300
301 SDGSSILDRA VFEHNLLSAS KLYNNITFEE LGALLDIPAV KAEKIASQMI TEGRMNGHID 360
361 QISAIVHFEN RELLPQWDRQ IQSLCYQVNS IIEKISVAEP DWMDNLN |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
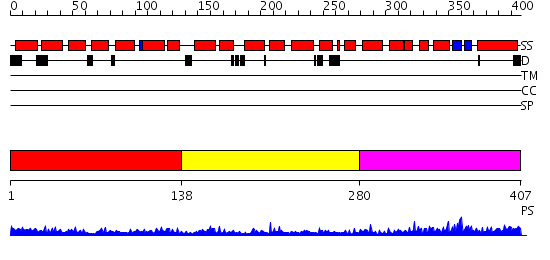
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..137] | 4.377996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [138..279] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [280..407] | 11.0 | Solution structure of the PCI domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)