













| General Information: |
|
| Name(s) found: |
PRRX1_RAT
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Rattus norvegicus |
| Length: | 245 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[UNKNOWN][IEA]
|
| Biological Process: |
multicellular organismal development
[IEA]
palate development [UNKNOWN][IEA] regulation of transcription, DNA-dependent [IEA] middle ear morphogenesis [UNKNOWN][IEA] embryonic cranial skeleton morphogenesis [UNKNOWN][IEA] artery morphogenesis [UNKNOWN][IEA] positive regulation of mesenchymal cell proliferation [UNKNOWN][IEA] embryonic skeletal system morphogenesis [UNKNOWN] negative regulation of transcription from RNA polymerase II promoter [IDA] embryonic limb morphogenesis [UNKNOWN][IEA] inner ear morphogenesis [UNKNOWN][IEA] positive regulation of smoothened signaling pathway [UNKNOWN][IEA] cartilage development [UNKNOWN][IEA] |
| Molecular Function: |
DNA binding
[UNKNOWN]
transcription factor activity [IEA] sequence-specific DNA binding [IEA] transcription repressor activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTSSYGHVLE RQPALGGRLD SPGNLDTLQA KKNFSVSHLL DLEEAGDMVA AQADESVGEA 60
61 GRSLLESPGL TSGSDTPQQD NDQLNSEEKK KRKQRRNRTT FNSSQLQALE RVFERTHYPD 120
121 AFVREDLARR VNLTEARVQV WFQNRRAKFR RNERAMLANK NASLLKSYSG DVTAVEQPIV 180
181 PRPAPRPTDY LSWGTASPYS AMATYSATCA NNSPAQGINM ANSIANLRLK AKEYSLQRNQ 240
241 VPTVN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
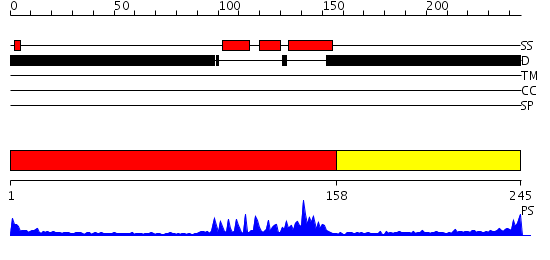
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..157] | 23.69897 | Paired protein | |
| 2 | View Details | [158..245] | 2.450995 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)