













| General Information: |
|
| Name(s) found: |
gi|119352249
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Plasmodium falciparum |
| Length: | 875 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
RNA processing
[ISS |
| Molecular Function: |
poly(A) RNA binding
[ISS mRNA binding [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIATGTNMMH PSFSTASLYV GDLNEDVTEA VLYEIFNTVG HVSSIRVCRD SVTRKSLGYA 60
61 YVNYHNLADA ERALDTLNYT NIKGQPARLM WSHRDPSLRK SGTGNIFVKN LDKSIDNKAL 120
121 FDTFSMFGNI LSCKVATDEF GKSKSYGFVH YEDEESAKEA IEKVNGVQLG SKNVYVGPFI 180
181 KKSERATNDT KFTNLYVKNF PDSVTETHLR QLFNPYGEIT SMIVKMDNKN RKFCFINYAD 240
241 AESAKNAMDN LNGKKITDDG QIDETYDPKK EEATASTSGA ANQTTGTDAD KTDKNDKNNK 300
301 ADKNEKGDSS NANNNATATG ATTTDTTTTP GETTTTTANA DSTGANNNNG LSPNTNTSNT 360
361 TTGSSNNSIN LNENNNTAGN NNSTNNNNNS GSSMNNAGSA KKDETAASDC ADTPNILYVG 420
421 PHQSRARRHA ILKAKFDNLN VENKNKHQGV NLYIKNLDDG IDDIMLRELF EPFGTITSAK 480
481 VMRDEKEQSK GFGFVCFASQ EEANKAVTEM HLKIINGKPL YVGLAEKREQ RLSRLQQRFR 540
541 MHPIRHHMNN PLNTPMQYAS PQSPQLQFSQ NTLSYGRPVI TAFNQNNLIS WRHQQAAQQQ 600
601 AVHQQAVHQQ AAQQQLNFNT NLRGQINQMR LYTQNNMMNN NLNQNKPNAQ LHHNQQYVPN 660
661 ALAQNGQQQP NLNAAGQHNA QQLQQQGNNQ LLNNNMRNMN NRANRNMGNL GNMNNQKQLP 720
721 LNINNKQQNA ASQANQMNHQ AQPQGAQAQQ KNPQQMQQVP QGNNFKFTAQ ARNRMELPNK 780
781 NANKVNTMNN NMNVNFNNNS TLTAAALASA PPSMQKQVLG ENLFPLVANY HPTLAGKITG 840
841 MMLEMDNSEL LILLENEEQL KKKIDEALVV LQKAK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
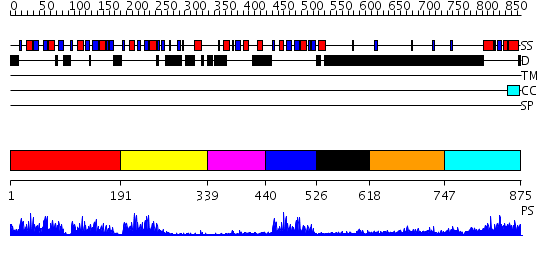
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..190] | 59.045757 | Poly(A)-binding protein | |
| 2 | View Details | [191..338] | 5.39794 | NMR Structure of RRM2 from Human Polypyrimidine Tract Binding Protein Isoform 1 (PTB1) | |
| 3 | View Details | [339..439] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [440..525] | 23.0 | No description for 2d9pA was found. | |
| 5 | View Details | [526..617] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [618..746] | 6.69897 | Sec24 | |
| 7 | View Details | [747..875] | 35.0 | poly(A) binding protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 7 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)