













| General Information: |
|
| Name(s) found: |
NTF3_MOUSE
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Mus musculus |
| Length: | 258 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum lumen
[UNKNOWN]
cytoplasmic membrane-bounded vesicle [IDA] extracellular region [UNKNOWN] plasma membrane [UNKNOWN] |
| Biological Process: |
neuromuscular synaptic transmission
[IDA]
epidermis development [IMP] nervous system development [UNKNOWN] positive regulation of glial cell differentiation [UNKNOWN] axon guidance [IDA][IMP] peripheral nervous system development [IMP] myelination [UNKNOWN] brain development [IMP] neuron development [IMP] generation of neurons [IGI] glial cell fate determination [IMP] positive regulation of transcription from RNA polymerase II promoter [IDA] regulation of neuron apoptosis [IDA] nerve development [IMP] enteric nervous system development [IMP] smooth muscle cell differentiation [IMP] mechanoreceptor differentiation [IGI] regulation of synaptic transmission [UNKNOWN] |
| Molecular Function: |
growth factor activity
[IEA]
protein binding [IPI] neurotrophin receptor binding [IEA] neurotrophin p75 receptor binding [TAS] receptor binding [IEA] nerve growth factor binding [UNKNOWN] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSILFYVIFL AYLRGIQGNS MDQRSLPEDS LNSLIIKLIQ ADILKNKLSK QMVDVKENYQ 60
61 STLPKAEAPR EPEQGEATRS EFQPMIATDT ELLRQQRRYN SPRVLLSDST PLEPPPLYLM 120
121 EDYVGNPVVA NRTSPRRKRY AEHKSHRGEY SVCDSESLWV TDKSSAIDIR GHQVTVLGEI 180
181 KTGNSPVKQY FYETRCKEAR PVKNGCRGID DKHWNSQCKT SQTYVRALTS ENNKLVGWRW 240
241 IRIDTSCVCA LSRKIGRT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
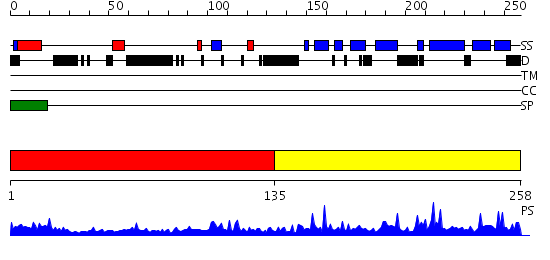
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..134] | 3.055976 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [135..258] | 56.0 | Neurotrophin 3, NT3 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.62 |
Source: Reynolds et al. (2008)