













| General Information: |
|
| Name(s) found: |
COG3_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 735 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFDQLEFYPA VNYEDNETQN DIKLPEVAKL ENIETVHSIS KERRDSLTEI LNDSSSLPAR 60
61 PFSLPNPNNS TVEKQSLFPF EEKMNPIWII SRPTFSIEQD AEKKINELQT FTNIIDQILG 120
121 QTNNIESTLL SMKEKFESSE KKLSEFSEMC ENLSTDEMRF SEIADGIRKG LTIFAPLKEL 180
181 TRVFRHPPPD FAGKVSFKEH ITQLNTCIMF LEENLDFQES PHYLGQYKKL LSQAMDIFKP 240
241 YFIRIIKQTT DQVLKDSKKM DVHKQLHSSL FYARFSAVGH NLCPTITELC KLCSKESLDA 300
301 FLPAFYDVYF QCRTRLLKPV LDYHLKSFFM EKSISSYIQK SLALLQLTFF DENKLFREIL 360
361 IMDDFRFMHY WNNLCQSFFE NSRSLILHEK NLTELCEVCS YIQSFQNAIL EGEREDVDKK 420
421 VVEFLNPLVL ELQERLLFVV QTAIETDIQR YSPTEEDLNP IADDKSLLFD LEKLRLNNDE 480
481 EKLDPDVPQK LAMAQGWYPV VQKSLIILSK IYRLVNSQVF DEIALELVHS CIRSLVDAYR 540
541 YFSRNNDKQL ARLFLIKNFL VLKDQLNSFD IHYACIEAGV DLRKVWDSVR EWRSNLRGVL 600
601 QLVYETFPKF ITNAVDTRQE LNQQLRVAVN GYIETAVIHY TECLSIGNLE SITEFQNRIQ 660
661 KFPELRQQIS LYLSETWIIE LFLQAIREEV VSKFQNFYES IVANEDITGS DHNKSGTTSL 720
721 KHLNEYVEDI YSVMS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
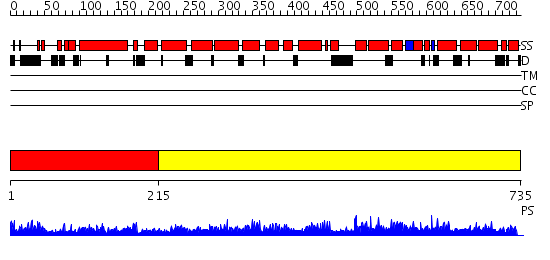
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..214] | 4.522879 | Crystal Structure Analysis of ClpB | |
| 2 | View Details | [215..735] | 1.23 | Crystal Structure of the S. cerevisiae Exocyst Component Exo70p |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)