













| General Information: |
|
| Name(s) found: |
KA111_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 990 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDGNEIAHAK KLVYDLYSGS LSPSAIAATE KELQKAQRSQ QGWNIGLFML QSKDVYEQFF 60
61 GALTLQMKIN TQLETLSDKD LVQLFVQLLQ KLLWDDGLPA LVERKVICTL ASLTIKYELE 120
121 KKEEASLKVI YCLFCNKLEF FNADMNAASV LNNLFPPASS RNAQLTASYI NELLLELSFS 180
181 IYTKENEDAL FNNVFRPCSN IIVSVLVFIF TNYSLDLSKE KNVAALEEAL NCMIAISSYL 240
241 AKASVSVQSV LPAFTECMDL TVNCIALDEV SEKAMNCLAD LLANYSNFIT QPTIERLWTI 300
301 LTGPWGETHL QQELEDPDSG EENDYSFLNI VIGFAEAMLP QIIDHIQEEK SIRLLYILAS 360
361 LLSFPGYAIV EEKVSWRTLE FWTTLIEDFS MSKAATDPSK DEIFKQIAFS VVEKAWWKML 420
421 LPSPEQWNSW PSSSRDSFNS YRRDLGDLLE SSYSIFGERL YAMYITTIEN FFSDGTGSPQ 480
481 SLEVSFYCLC CILEYDTNDS DTLDAWLTRL FETSFAIKAS AFQNPQLLKT CSQLLSSCSC 540
541 FLQNHPQYLN ISLPVLFDAL HISETSIQMT VSRSIHTLCT TCASHLLTEI DGFMAVVEEL 600
601 TPKLVYVPSV LEKIYSSVGY VTQRIEDIEL RISYLMRLLN CILAQLQPSL YPNLEIFENV 660
661 LKSCLQSVAG VALSQSPIGE SPIIDVEQST QETTFWQQSC IAEFQAKLIS FLTHSESMAL 720
721 QYSDVVGLIC KIMIAGLNEV EPSPFSLPIV TTIQYFCDRF TEFPAAVLLT LGSAILTCPY 780
781 GQTDIIDKVL IDMCSSIQNS VVIINEESFM NNIDITVELY HFFSIILQKH PSFLETMYPD 840
841 FTQLILNRAI NLLGKPERLL ESAAGQFIIS FITSEKSDLL NTHTDFVNAI RSPLIAKILL 900
901 GFGGNASRSS LPLLSDILGK LKAQNFSATR ACLTQSLEEE GFPSRNVSNE IKRRFLTDLL 960
961 KARIKDKVKQ FWILCKGLES TPYGNSSWTF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
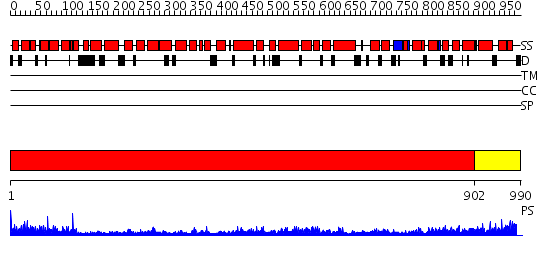
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..901] | 91.39794 | Karyopherin beta2 | |
| 2 | View Details | [902..990] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)