













| General Information: |
|
| Name(s) found: |
PNK1_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 421 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSKKRKSPP QESLTSYFEK SSKSSKKYGS QNKDSDSSST CLQQKIEIQW SITDSLYIAK 60
61 YGKLKKTKKF IAFDLDGTLI KTKSGRVFSK DAADWTWWHP SVVPKLKALY QDNYSLVIFS 120
121 NQNGIPRKPS AGHTFQMKIR AIFESLDLPI VLYAAILKDK FRKPLTGMWN SFLKDVNRSI 180
181 DLSFIKYVGD AAGRPGDHNS TDLKFAENIG IKFETPEQFF LGHSFVPPNF ESFHPKNYLV 240
241 RNSSSHPYHF KKSEHQEIVV LVGFPSSGKS TLAESQIVTQ GYERVNQDIL KTKSKCIKAA 300
301 IEALKKEKSV VIGMYSIIST TYAISDNTNP TIESRKMWID IAQEFEIPIR CIHLQSSEEL 360
361 ARHNNVFRYI HHNQKQLPEI AFNSFKSRFQ MPTVEEGFTN VEEVPFKCLK DYEDTWNYWY 420
421 E |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
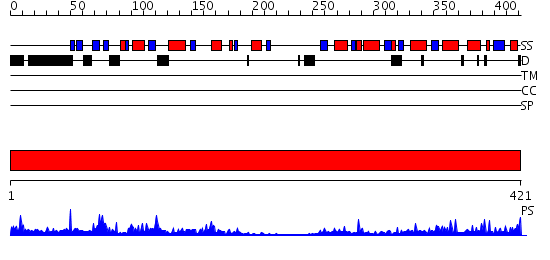
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..421] | 66.09691 | Molecular architecture of mammalian polynucleotide kinase, a DNA repair enzyme |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)