













| General Information: |
|
| Name(s) found: |
DIS3_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 970 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTVSGLKRP QSSEKNHRDR VFVRATRGKV QKVVREQYLR NDIPCQSRAC PLCRSKLPKD 60
61 SRGNVLEPIL SEKPMFLEKF GHHYLIPDSN IFYHCIDALE HPNNFFDVII LQTVFSEISS 120
121 KSIPLYNRMK RLCQEKTKRF TPFSNEFFVD TFVERLDDES ANDRNDRAIR NAASWFASHL 180
181 ASLGIKIVLL TDDRENARLA AEQGIQVSTL KDYVQYLPDS EILLDMVSAI ADAIASKEQV 240
241 ESGTKNVYEL HWSMSRLLAC IKNGEVHKGL INISTYNYLE GSVVVPGYNK PVLVSGRENL 300
301 NRAVQGDIVC IQILPQDQWK TEAEEIADDD EDVVVSTAAE PDSARINDLE LITKRNAHPT 360
361 AKVVGILKRN WRPYVGHVDN ATIAQSKGGS QQTVLLTPMD RRVPKIRFRT RQAPRLVGRR 420
421 IVVAIDLWDA SSRYPEGHFV RDLGEMETKE AETEALLLEY DVQHRPFPKA VLDCLPEEGH 480
481 NWKVPADKTH PLWKNRKDFR DKLICSIDPP GCQDIDDALH ACVLPNGNYE VGVHIADVTH 540
541 FVKPNTSMDS EAASRGTTVY LVDKRIDMLP MLLGTDLCSL RPYVERFAFS CIWEMDENAN 600
601 IIKVHFTKSV IASKEAFSYA DAQARIDDQK MQDPLTQGMR VLLKLSKILK QKRMDEGALN 660
661 LASPEVRIQT DNETSDPMDV EIKQLLETNS LVEEFMLLAN ISVAQKIYDA FPQTAVLRRH 720
721 AAPPLTNFDS LQDILRVCKG MHLKCDTSKS LAKSLDECVD PKEPYFNTLL RILTTRCMLS 780
781 AEYFCSGTFA PPDFRHYGLA SPIYTHFTSP IRRYADVLAH RQLAAAIDYE TINPSLSDKS 840
841 RLIEICNGIN YRHRMAQMAG RASIEYYVGQ ALKGGVAEED AYVIKVFKNG FVVFIARFGL 900
901 EGIVYTKSLS SVLEPNVEYV EDEYKLNIEI RDQPKPQTVQ IQMFQQVRVR VTTVRDEHSG 960
961 KQKVQITLVY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
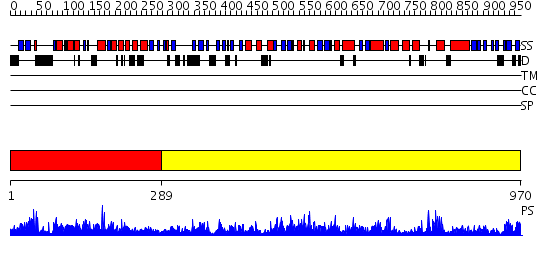
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..288] | 4.81 | No description for 2dokA was found. | |
| 2 | View Details | [289..970] | 138.0 | No description for 2ix0A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)