













| General Information: |
|
| Name(s) found: |
Nipped-B-PE /
FBpp0110411
[FlyBase]
Nipped-B-PF / FBpp0110410 [FlyBase] |
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 2077 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cohesin complex [IDA] protein complex [IDA] synaptonemal complex [IDA] |
| Biological Process: |
positive regulation of gene expression
[IGI][IMP]
mitotic sister chromatid cohesion [IMP] regulation of transcription, DNA-dependent [IMP] |
| Molecular Function: |
transcription activator activity
[IMP]
binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGERDNPTVP VTTLAGLTST SDLLSELPVA DSLQSAASLN KSLLFHALVA NESSNLLSMR 60
61 NENLVRQLVT AIERTNSDNI ELIHCPVQDT ATNCSTYPEL LQGIYHFRPA VFNTSIKIHS 120
121 PDQHTANARL EAKKMQIDSY SIPECYASPT NLSISNEHQL QDAQACFNQL TSMTSKEILE 180
181 EFSQINFKEN ATEKDKIDVI ENSDLNVTKI LSQNTLKKKS NTPERSTVQS IQEQFFIQQQ 240
241 EANYNLKHDL NQVSTNVGQI QNISDTFPQE PSNFNLNSVN IPNMLYVQSP PFTESEPTQA 300
301 HHSSIEYLKK KKSQILDVSI LNRQQVLENT SLHIINKSST YQTNQNVSTT TSTSTSSSGK 360
361 SQVRVCINRL SIEDSRLMQQ SIKKFVQKSP ELARSMGLLQ ETCQQQHENL NIIPTSAEEC 420
421 VLESRASSTN NTVKIPIDTF STIKADEKRS AKRKLAISIG DIPPEQIFSK PKMRRVERIT 480
481 PLSNTKCVKE EVTRSQTYQQ FIRNMDHIIE ILDDSESPNF DSEDVDETIE CISSKLLNSM 540
541 STDVAKLKAK QALDSIPKNK LTLLINYAMR NVYLARNYFA GTEDEDEFVD DEVIEKLLNA 600
601 MDACLLICNI YSTVSDLQFL QEDNVSHIIK FTQFQLRETI FPLHDPVYTA KSIKRTTHRK 660
661 KIKSHQAQNR SMQLFYLKTV ELLKVFVTLF DKCVFVDTIV LPLSTLAIEP FFVDNIETLQ 720
721 FVCLELVTTI FRKERYDKIR NSILGDILTS IDRLPSSKKN LRPYKLTNNG GNIQMVTALV 780
781 LQLIQCATIL PDSLCDNGKF SNKPQEGNTF DEEGKKLLQP SQDLLVLQKY DVAVSIGGNF 840
841 LTTFLNKCKS RSNETDFRPL FENFIHDLLA TVNKPEWPAS ELLLSLLGTM LVRYVSDKGI 900
901 EQSIRLVSLD YLGIVAARLR KDTVESRCRV NIIDSMIQSI KLEQEKEGDV TSNNDQFDLE 960
961 PEEQRTDFLQ KILLDFLAVN AQEENLIWDY ARHFYLAQWY RDVIYQRRRI NDGKKGLAFR1020
1021 KSKIRNNRRT NGDYLDTSDS GSCDESDTDT NKKRIHCVDS NDFELNINIY KALEARKQYF1080
1081 INKIKPFSVF GEQNHSSNQH IKTYIDYNNA QLIAQYLATK RPFSQSFDGC LKKIILVVNE1140
1141 PSIAVRTRAM KCLANIVEVD PLVLKRKDMQ MGVNQKFLDT AISVREAAVD LVGKFVLSNQ1200
1201 DLIDQYYDML STRILDTGVS VRKRVIKILR DICLEYPDFS KIPEICVKMI RRVHDEEGIQ1260
1261 KLVTEVFMKM WFTPCTKNDK IGIQRKINHI IDVVNTAHDT GTTWLEGLLM SIFKPRDNML1320
1321 RSEGCVQEFI KKNSEPPMDI VIACQQLADG LVDRLIELED TDNSRMLGCI TTLHLLAKVR1380
1381 PQLLVKHAIT IEPYLNIKCH SATAAKFICA VADILEKVVP LVNNASESFL ASLEEHLMLL1440
1441 VVSRNQAEVT SCVSCLGALV NKITHNFKLI RDCFQKFYRV LDVSRSQVIQ GNNSVDNIYT1500
1501 PSFRRSLFTI GILMRYFDFK SPIALGETND GLPVSICEDV FHCLMFFCRC TNQEIRKQAL1560
1561 ISLGSFCVLN DGYLTRSELK NLYCEILSSI ANDAGFKIIC MRNIWIYLTE SEMFMHNKEK1620
1621 EWEKQSKHED LKEMNDVSSG MASRIIQLYL EEILECFLNR DDTVRLWAVK VIQIVLRQGL1680
1681 VHPVRMVPYL ICLSTDHRIE SAHRADALLK DIDKTYSGFV NMKVQFGLQL CFKLQKILQI1740
1741 NNRGKLEIIR GYASRGPDNT TTALNDFLYT LLRTTKPQRR ALVQTVTKQF DDQKTSLQQM1800
1801 LYIADNLAYF PYVVQDEPLY LIHQIDLLIS MAGTHLLATF KEHIKPSDKE GDVLEDDDDV1860
1861 EDPEVLFNRL PEDLTEIIKC ITSAQACMLL LILKDHLKEM YAITDSKISR YSPSEQKLYE1920
1921 KAVTRKSVND FNPKTTIDVI KKQMSQEKLS TDINFTLTKE EKLDLVVKYL DFKQLMLKLD1980
1981 PDDGDSDADE SRDKTMLNIS ASSDGVAFSS SAKNSHSACD GYSLTVTDVV DVPMSHIAKA2040
2041 SMLTSKPSGR KTNPVRTKKK RRKIDSTDDE TSDAEYA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
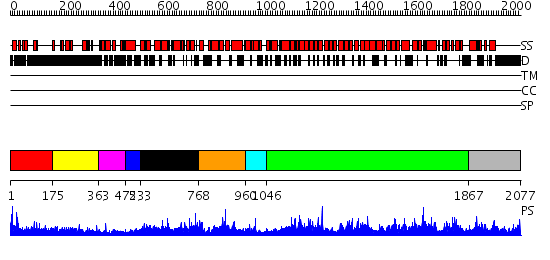
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..174] | 1.405991 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [175..362] | 6.69897 | Sec24 | |
| 3 | View Details | [363..471] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [472..532] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [533..767] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [768..959] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [960..1045] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1046..1866] | 2.23 | Adaptin beta subunit N-terminal fragment | |
| 9 | View Details | [1867..2077] | 1.047961 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||
| 8 |
|
||||||||||||||||||||||||||||||
| 9 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)