













| General Information: |
|
| Name(s) found: |
AP1B1
[HGNC (HUGO)]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 949 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTDSKYFTTT KKGEIFELKA ELNSDKKEKK KEAVKKVIAS MTVGKDVSAL FPDVVNCMQT 60
61 DNLELKKLVY LYLMNYAKSQ PDMAIMAVNT FVKDCEDPNP LIRALAVRTM GCIRVDKITE 120
121 YLCEPLRKCL KDEDPYVRKT AAVCVAKLHD INAQLVEDQG FLDTLKDLIS DSNPMVVANA 180
181 VAALSEIAES HPSSNLLDLN PQSINKLLTA LNECTEWGQI FILDCLANYM PKDDREAQSI 240
241 CERVTPRLSH ANSAVVLSAV KVLMKFMEML SKDLDYYGTL LKKLAPPLVT LLSAEPELQY 300
301 VALRNINLIV QKRPEILKHE MKVFFVKYND PIYVKLEKLD IMIRLASQAN IAQVLAELKE 360
361 YATEVDVDFV RKAVRAIGRC AIKVEQSAER CVSTLLDLIQ TKVNYVVQEA IVVIKDIFRK 420
421 YPNKYESVIA TLCENLDSLD EPEARAAMIW IVGEYAERID NADELLESFL EGFHDESTQV 480
481 QLQLLTAIVK LFLKKPTETQ ELVQQVLSLA TQDSDNPDLR DRGYIYWRLL STDPVAAKEV 540
541 VLAEKPLISE ETDLIEPTLL DELICYIGTL ASVYHKPPSA FVEGGRGVVH KSLPPRTASS 600
601 ESAESPETAP TGAPPGEQPD VIPAQGDLLG DLLNLDLGPP VSGPPLATSS VQMGAVDLLG 660
661 GGLDSLMGDE PEGIGGTNFV APPTAAVPAN LGAPIGSGLS DLFDLTSGVG TLSGSYVAPK 720
721 AVWLPAMKAK GLEISGTFTR QVGSISMDLQ LTNKALQVMT DFAIQFNRNS FGLAPATPLQ 780
781 VHAPLSPNQT VEISLPLSTV GSVMKMEPLN NLQVAVKNNI DVFYFSTLYP LHILFVEDGK 840
841 MDRQMFLATW KDIPNENEAQ FQIRDCPLNA EAASSKLQSS NIFTVAKRNV EGQDMLYQSL 900
901 KLTNGIWVLA ELRIQPGNPS CTDLELSLKC RAPEVSQHVY QAYETILKN |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
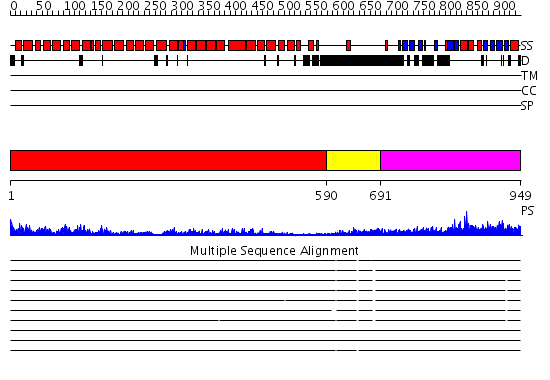
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..589] | 104.0 | Adaptin beta subunit N-terminal fragment | |
| 2 | View Details | [590..690] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [691..949] | 70.154902 | Beta2-adaptin AP2 ear domain, N-terminal subdomain; Beta2-adaptin AP2, C-terminal subdomain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)