













| General Information: |
|
| Name(s) found: |
gi|108864074
[NCBI NR]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 651 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPRKASSTSD SRLKWRKWKR NPTASPSPSN RSSAAAAAAD HSDDSDSAAV NEDDDSAVPE 60
61 DADDETLAGA EDPVLDLREA EVLPSAEPVS AFPVATRRVV NRPHPSVLAV IAAERSACAG 120
121 EGSAAVAAAP VLENISYGQQ QVLSGVLPDH ASLATDTDKP STYVCTPPNL MEGHGVTKQF 180
181 QGRLHVVPKH SDWFSPGIVH RLERQVVPQF FSGKSPGNTP EKYMLLRNKV IAKYLENPSK 240
241 RLAFAECQGL VANTAELYDL SRIVRFLDTW GIINYLASGS VHRGLRMATS LLREEPTGEL 300
301 QLLTAPLKSI DGLILFDRPK CNLQAEDISS LASNSEVVDF DAGLAELDGK IRERLSESSC 360
361 SYCLQPLTSL HYQSLKEADI ALCSDCFHDA RYITGHSSLD FQRIDGDNDR SENDGDSWTD 420
421 QETLLLLEGI EKYNDNWNNI AEHVGTKSKA QCIYHFIRLP VEDGLLENIE VPDASVPFRA 480
481 ETNGYPHLDC NGSTSGNLPQ KIPPDNQLPF INSSNPVMSL VGFLASAMGP RVAASCASAA 540
541 LSVLTVDDDS RVNSEGICSD SRGQGPHPNF RDHNGGVSSS ISPEKVKHAA MCGLSAAATK 600
601 AKLFADQEER EIQRLTATVI NHQVSVNVAI LFTFGPCFSH YCCHQGVKNR R |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
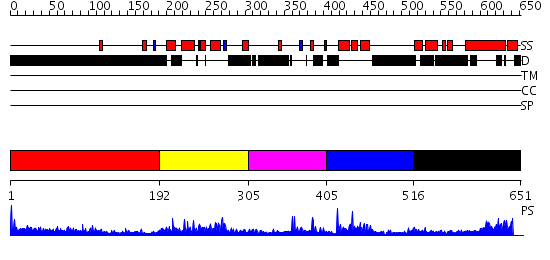
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..191] | 24.175996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [192..304] | 27.154902 | Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes | |
| 3 | View Details | [305..404] | 2.35 | No description for 2fc7A was found. | |
| 4 | View Details | [405..515] | 7.39794 | No description for 2iw5B was found. | |
| 5 | View Details | [516..651] | 1.12499 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|