













| General Information: |
|
| Name(s) found: |
gi|108706704
[NCBI NR]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 586 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGKKQHSKDR MFITRTEWAT EWGGAKQKEA GTPFKRLPFY CCALTFLPFE DPVCTADGSV 60
61 FDLMSIIPYI KKFGKHPVTG TPLKQEDLMP LTFHKNSDGE FQCPVLNKVF TEFTHIVAVK 120
121 TTGNVFCYEA IQELNIKPKN WRELLTDEPF TRNDLITIQN PNAVDSKILG EFDHVKKGLK 180
181 LEDEELQRMK NDPTYNINIS GDLKQMIKEL GTEKGKLAFL HGGGGQKAQK ERAAALAAIL 240
241 AKKEKDDSKS GKEPKPHQPF SIVDAASASV HGRSAAAAKA ATAEKTAARI AMHMAGDRAP 300
301 VNAKLVKSRY TTGAASRSFT STAYDPVTKN ELEYVKVEKN PKKKGYVQLH TTHGDLNLEL 360
361 HCDITPRTCE NFLTHCENGY YNGLIFHRSI KNFMIQGGDP TGTGSGGESI WGKPFKDELN 420
421 SKLIHSGRGV VSMANSGPHT NGSQFFILYK SAPHLNFKHT VFGMVVGGLT TLSAMEKVPV 480
481 DDDDRPLEEI KILKVSVFVN PYTEPDEEEE EKAKEEEKKK DEDYDKVGSW YSNPGTGVAG 540
541 STSSGGGVGK YLKARTAGFA DVVADDSNKK RKASVSNVEF KDFSGW |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
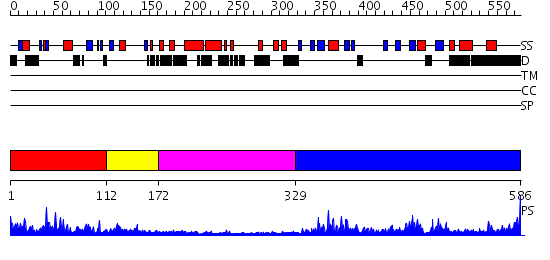
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..111] | 13.30103 | No description for 2f42A was found. | |
| 2 | View Details | [112..171] | 2.24 | Not-4 N-terminal RING finger domain | |
| 3 | View Details | [172..328] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [329..586] | 51.30103 | Crystal Structure of the cyclophiln_RING domain of human peptidylprolyl isomerase (cyclophilin)-like 2 isoform b |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)