













| General Information: |
|
| Name(s) found: |
PRKAG2
[HGNC (HUGO)]
|
| Description(s) found:
Found 33 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 569 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSAVMDTKK KKDVSSPGGS GGKKNASQKR RSLRVHIPDL SSFAMPLLDG DLEGSGKHSS 60
61 RKVDSPFGPG SPSKGFFSRG PQPRPSSPMS APVRPKTSPG SPKTVFPFSY QESPPRSPRR 120
121 MSFSGIFRSS SKESSPNSNP ATSPGGIRFF SRSRKTSGLS SSPSTPTQVT KQHTFPLESY 180
181 KHEPERLENR IYASSSPPDT GQRFCPSSFQ SPTRPPLASP THYAPSKAAA LAAALGPAEA 240
241 GMLEKLEFED EAVEDSESGV YMRFMRSHKC YDIVPTSSKL VVFDTTLQVK KAFFALVANG 300
301 VRAAPLWESK KQSFVGMLTI TDFINILHRY YKSPMVQIYE LEEHKIETWR ELYLQETFKP 360
361 LVNISPDASL FDAVYSLIKN KIHRLPVIDP ISGNALYILT HKRILKFLQL FMSDMPKPAF 420
421 MKQNLDELGI GTYHNIAFIH PDTPIIKALN IFVERRISAL PVVDESGKVV DIYSKFDVIN 480
481 LAAEKTYNNL DITVTQALQH RSQYFEGVVK CNKLEILETI VDRIVRAEVH RLVVVNEADS 540
541 IVGIISLSDI LQALILTPAG AKQKETETE |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
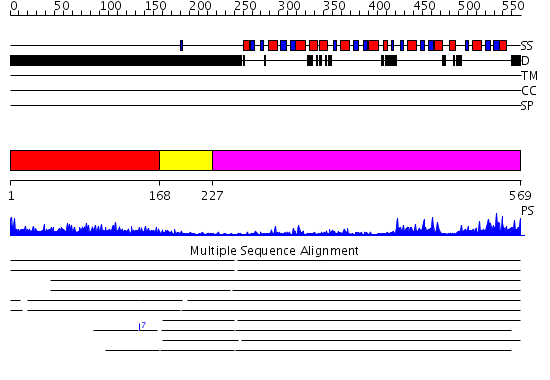
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..167] | 1.34 | No description for 2j63A was found. | |
| 2 | View Details | [168..226] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [227..569] | 48.69897 | No description for 2v8qE was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)