













| General Information: |
|
| Name(s) found: |
GAK
[HGNC (HUGO)]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 1311 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLLQSALDF LAGPGSLGGA SGRDQSDFVG QTVELGELRL RVRRVLAEGG FAFVYEAQDV 60
61 GSGREYALKR LLSNEEEKNR AIIQEVCFMK KLSGHPNIVQ FCSAASIGKE ESDTGQAEFL 120
121 LLTELCKGQL VEFLKKMESR GPLSCDTVLK IFYQTCRAVQ HMHRQKPPII HRDLKVENLL 180
181 LSNQGTIKLC DFGSATTISH YPDYSWSAQR RALVEEEITR NTTPMYRTPE IIDLYSNFPI 240
241 GEKQDIWALG CILYLLCFRQ HPFEDGAKLR IVNGKYSIPP HDTQYTVFHS LIRAMLQVNP 300
301 EERLSIAEVV HQLQEIAAAR NVNPKSPITE LLEQNGGYGS ATLSRGPPPP VGPAGSGYSG 360
361 GLALAEYDQP YGGFLDILRG GTERLFTNLK DTSSKVIQSV ANYAKGDLDI SYITSRIAVM 420
421 SFPAEGVESA LKNNIEDVRL FLDSKHPGHY AVYNLSPRTY RPSRFHNRVS ECGWAARRAP 480
481 HLHTLYNICR NMHAWLRQDH KNVCVVHCMD GRAASAVAVC SFLCFCRLFS TAEAAVYMFS 540
541 MKRCPPGIWP SHKRYIEYMC DMVAEEPITP HSKPILVRAV VMTPVPLFSK QRSGCRPFCE 600
601 VYVGDERVAS TSQEYDKMRD FKIEDGKAVI PLGVTVQGDV LIVIYHARST LGGRLQAKMA 660
661 SMKMFQIQFH TGFVPRNATT VKFAKYDLDA CDIQEKYPDL FQVNLEVEVE PRDRPSREAP 720
721 PWENSSMRGL NPKILFSSRE EQQDILSKFG KPELPRQPGS TAQYDAGAGS PEAEPTDSDS 780
781 PPSSSADASR FLHTLDWQEE KEAETGAENA SSKESESALM EDRDESEVSD EGGSPISSEG 840
841 QEPRADPEPP GLAAGLVQQD LVFEVETPAV LPEPVPQEDG VDLLGLHSEV GAGPAVPPQA 900
901 CKAPSSNTDL LSCLLGPPEA ASQGPPEDLL SEDPLLLASP APPLSVQSTP RGGPPAAADP 960
961 FGPLLPSSGN NSQPCSNPDL FGEFLNSDSV TVPPSFPSAH SAPPPSCSAD FLHLGDLPGE1020
1021 PSKMTASSSN PDLLGGWAAW TETAASAVAP TPATEGPLFS PGGQPAPCGS QASWTKSQNP1080
1081 DPFADLGDLS SGLQGSPAGF PPGGFIPKTA TTPKGSSSWQ TSRPPAQGAS WPPQAKPPPK1140
1141 ACTQPRPNYA SNFSVIGARE ERGVRAPSFA QKPKVSENDF EDLLSNQGFS SRSDKKGPKT1200
1201 IAEMRKQDLA KDTDPLKLKL LDWIEGKERN IRALLSTLHT VLWDGESRWT PVGMADLVAP1260
1261 EQVKKHYRRA VLAVHPDKAA GQPYEQHAKM IFMELNDAWS EFENQGSRPL F |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
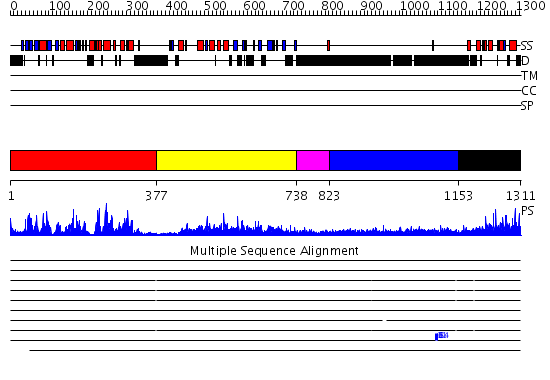
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..376] | 83.0 | No description for 2qnjA was found. | |
| 2 | View Details | [377..737] | 81.0 | Pten tumor suppressor (Phoshphoinositide phosphatase), C-terminal domain; Phoshphoinositide phosphatase Pten (Pten tumor suppressor), N-terminal domain | |
| 3 | View Details | [738..822] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [823..1152] | 4.221849 | No description for 1y0fA was found. | |
| 5 | View Details | [1153..1311] | 66.522879 | NMR Structure of the J-Domain and Clathrin Substrate Binding Domain of Bovine Auxilin |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|