













| General Information: |
|
| Name(s) found: |
GABBR1
[HGNC (HUGO)]
|
| Description(s) found:
Found 46 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 961 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLLLLLLAPL FLRPPGAGGA QTPNATSEGC QIIHPPWEGG IRYRGLTRDQ VKAINFLPVD 60
61 YEIEYVCRGE REVVGPKVRK CLANGSWTDM DTPSRCVRIC SKSYLTLENG KVFLTGGDLP 120
121 ALDGARVDFR CDPDFHLVGS SRSICSQGQW STPKPHCQVN RTPHSERRAV YIGALFPMSG 180
181 GWPGGQACQP AVEMALEDVN SRRDILPDYE LKLIHHDSKC DPGQATKYLY ELLYNDPIKI 240
241 ILMPGCSSVS TLVAEAARMW NLIVLSYGSS SPALSNRQRF PTFFRTHPSA TLHNPTRVKL 300
301 FEKWGWKKIA TIQQTTEVFT STLDDLEERV KEAGIEITFR QSFFSDPAVP VKNLKRQDAR 360
361 IIVGLFYETE ARKVFCEVYK ERLFGKKYVW FLIGWYADNW FKIYDPSINC TVDEMTEAVE 420
421 GHITTEIVML NPANTRSISN MTSQEFVEKL TKRLKRHPEE TGGFQEAPLA YDAIWALALA 480
481 LNKTSGGGGR SGVRLEDFNY NNQTITDQIY RAMNSSSFEG VSGHVVFDAS GSRMAWTLIE 540
541 QLQGGSYKKI GYYDSTKDDL SWSKTDKWIG GSPPADQTLV IKTFRFLSQK LFISVSVLSS 600
601 LGIVLAVVCL SFNIYNSHVR YIQNSQPNLN NLTAVGCSLA LAAVFPLGLD GYHIGRNQFP 660
661 FVCQARLWLL GLGFSLGYGS MFTKIWWVHT VFTKKEEKKE WRKTLEPWKL YATVGLLVGM 720
721 DVLTLAIWQI VDPLHRTIET FAKEEPKEDI DVSILPQLEH CSSRKMNTWL GIFYGYKGLL 780
781 LLLGIFLAYE TKSVSTEKIN DHRAVGMAIY NVAVLCLITA PVTMILSSQQ DAAFAFASLA 840
841 IVFSSYITLV VLFVPKMRRL ITRGEWQSEA QDTMKTGSST NNNEEEKSRL LEKENRELEK 900
901 IIAEKEERVS ELRHQLQSRQ QLRSRRHPPT PPEPSGGLPR GPPEPPDRLS CDGSRVHLLY 960
961 K |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
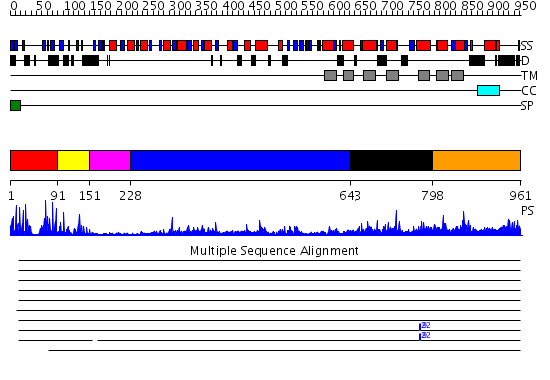
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..90] | 1.06 | No description for 2ehfA was found. | |
| 2 | View Details | [91..150] | 4.30103 | Solution structure of the second complement control protein (CCP) module of the GABA(B)R1a receptor, Pro-119 trans conformer | |
| 3 | View Details | [151..227] | 2.94 | AMIDE RECEPTOR/NEGATIVE REGULATOR OF THE AMIDASE OPERON OF PSEUDOMONAS AERUGINOSA (AMIC) COMPLEXED WITH ACETAMIDE | |
| 4 | View Details | [228..642] | 36.522879 | No description for 2e4uA was found. | |
| 5 | View Details | [643..797] | 6.479975 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [798..961] | 4.69897 | N-methyl-D-aspartate receptor subunit 1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|