













| General Information: |
|
| Name(s) found: |
HSF1
[HGNC (HUGO)]
|
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 529 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDLPVGPGAA GPSNVPAFLT KLWTLVSDPD TDALICWSPS GNSFHVFDQG QFAKEVLPKY 60
61 FKHNNMASFV RQLNMYGFRK VVHIEQGGLV KPERDDTEFQ HPCFLRGQEQ LLENIKRKVT 120
121 SVSTLKSEDI KIRQDSVTKL LTDVQLMKGK QECMDSKLLA MKHENEALWR EVASLRQKHA 180
181 QQQKVVNKLI QFLISLVQSN RILGVKRKIP LMLNDSGSAH SMPKYSRQFS LEHVHGSGPY 240
241 SAPSPAYSSS SLYAPDAVAS SGPIISDITE LAPASPMASP GGSIDERPLS SSPLVRVKEE 300
301 PPSPPQSPRV EEASPGRPSS VDTLLSPTAL IDSILRESEP APASVTALTD ARGHTDTEGR 360
361 PPSPPPTSTP EKCLSVACLD KNELSDHLDA MDSNLDNLQT MLSSHGFSVD TSALLDLFSP 420
421 SVTVPDMSLP DLDSSLASIQ ELLSPQEPPR PPEAENSSPD SGKQLVHYTA QPLFLLDPGS 480
481 VDTGSNDLPV LFELGEGSYF SEGDGFAEDP TISLLTGSEP PKAKDPTVS |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
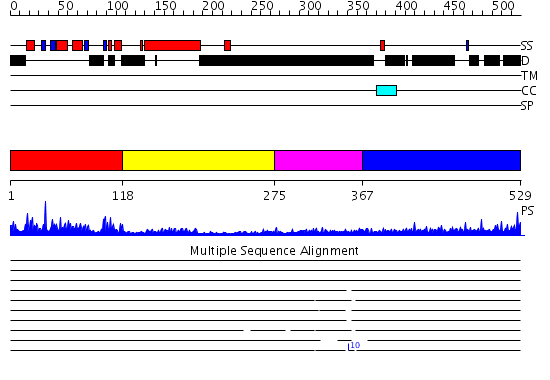
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..117] | 37.69897 | SOLUTION STRUCTURE OF THE DNA-BINDING DOMAIN OF DROSOPHILA HEAT SHOCK TRANSCRIPTION FACTOR | |
| 2 | View Details | [118..274] | 9.721246 | No description for PF00447.8 was found. No confident structure predictions are available. | |
| 3 | View Details | [275..366] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [367..529] | 7.145997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)