













| General Information: |
|
| Name(s) found: |
BRF1
[HGNC (HUGO)]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 677 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTGRVCRGCG GTDIELDAAR GDAVCTACGS VLEDNIIVSE VQFVESSGGG SSAVGQFVSL 60
61 DGAGKTPTLG GGFHVNLGKE SRAQTLQNGR RHIHHLGNQL QLNQHCLDTA FNFFKMAVSR 120
121 HLTRGRKMAH VIAACLYLVC RTEGTPHMLL DLSDLLQVNV YVLGKTFLLL ARELCINAPA 180
181 IDPCLYIPRF AHLLEFGEKN HEVSMTALRL LQRMKRDWMH TGRRPSGLCG AALLVAARMH 240
241 DFRRTVKEVI SVVKVCESTL RKRLTEFEDT PTSQLTIDEF MKIDLEEECD PPSYTAGQRK 300
301 LRMKQLEQVL SKKLEEVEGE ISSYQDAIEI ELENSRPKAK GGLASLAKDG STEDTASSLC 360
361 GEEDTEDEEL EAAASHLNKD LYRELLGGAP GSSEAAGSPE WGGRPPALGS LLDPLPTAAS 420
421 LGISDSIREC ISSQSSDPKD ASGDGELDLS GIDDLEIDRY ILNESEARVK AELWMRENAE 480
481 YLREQREKEA RIAKEKELGI YKEHKPKKSC KRREPIQAST AREAIEKMLE QKKISSKINY 540
541 SVLRGLSSAG GGSPHREDAQ PEHSASARKL SRRRTPASRS GADPVTSVGK RLRPLVSTQP 600
601 AKKVATGEAL LPSSPTLGAE PARPQAVLVE SGPVSYHADE EADEEEPDEE DGEPCVSALQ 660
661 MMGSNDYGCD GDEDDGY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
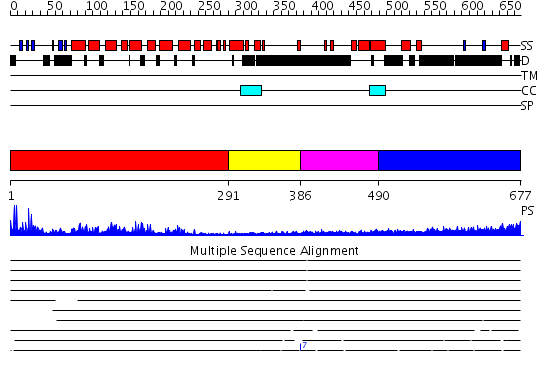
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..290] | 37.154902 | No description for 2g9xB was found. | |
| 2 | View Details | [291..385] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [386..489] | 2.69 | TBP-binding domain of the transcription factor IIIb Brf1 subunit | |
| 4 | View Details | [490..677] | 4.235996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)