













| General Information: |
|
| Name(s) found: |
CYC2 /
YOR037W
[SGD]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 366 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA extrinsic to mitochondrial inner membrane [IDA |
| Biological Process: |
cytochrome c-heme linkage
[IGI mitochondrial membrane organization [IMP |
| Molecular Function: |
oxidoreductase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLWKNYVLSS SRITRRLHKS PRKSSFSKNF FITGCLLTVG AVSSYLTYRY TSERENKHEL 60
61 SPSYFVKYKI SHKRDIDSSH FLLEVTPLFK QKVNIWSLMT AENLWSVEIK QPEVMVVRNY 120
121 TPLPLKFNPA SKEIEILKDG DNADGKLSFY IKKYENGEVA RWLHHLPKGH IIEIRGPFID 180
181 YEFPHLPNEL KRSRDCLYMD NRNERGNNVR ENSQFIYQPY DIMMFTAGTG IVTALQLLLT 240
241 ESPFRGTIKL FHTDKNIKQL GPLYPILLRL QASNRVQLKI FETDRQTKQD VLKSIQKSIT 300
301 KPYPYKGLLP FSNVNNKNIM PVLALVCGPE SYISSISGRK YDLNQGPVGG LLSKEGWNSD 360
361 NVYKLS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |

|
CYC2 QCR7 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
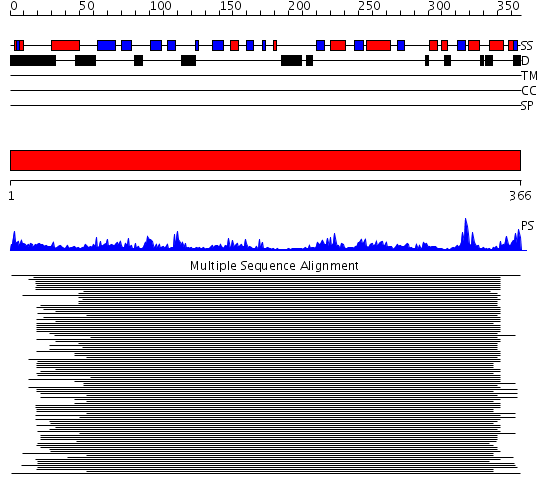
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..366] | 51.0 | cytochrome b5 reductase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|