













| General Information: |
|
| Name(s) found: |
INO1 /
YJL153C
[SGD]
|
| Description(s) found:
Found 56 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 533 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
inositol metabolic process
[TAS |
| Molecular Function: |
inositol-3-phosphate synthase activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTEDNIAPIT SVKVVTDKCT YKDNELLTKY SYENAVVTKT ASGRFDVTPT VQDYVFKLDL 60
61 KKPEKLGIML IGLGGNNGST LVASVLANKH NVEFQTKEGV KQPNYFGSMT QCSTLKLGID 120
121 AEGNDVYAPF NSLLPMVSPN DFVVSGWDIN NADLYEAMQR SQVLEYDLQQ RLKAKMSLVK 180
181 PLPSIYYPDF IAANQDERAN NCINLDEKGN VTTRGKWTHL QRIRRDIQNF KEENALDKVI 240
241 VLWTANTERY VEVSPGVNDT MENLLQSIKN DHEEIAPSTI FAAASILEGV PYINGSPQNT 300
301 FVPGLVQLAE HEGTFIAGDD LKSGQTKLKS VLAQFLVDAG IKPVSIASYN HLGNNDGYNL 360
361 SAPKQFRSKE ISKSSVIDDI IASNDILYND KLGKKVDHCI VIKYMKPVGD SKVAMDEYYS 420
421 ELMLGGHNRI SIHNVCEDSL LATPLIIDLL VMTEFCTRVS YKKVDPVKED AGKFENFYPV 480
481 LTFLSYWLKA PLTRPGFHPV NGLNKQRTAL ENFLRLLIGL PSQNELRFEE RLL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
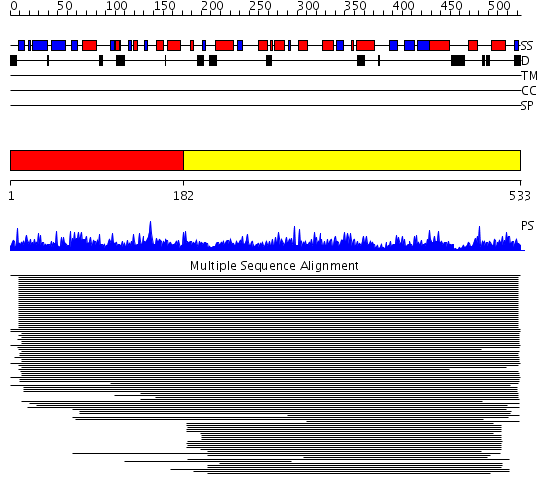
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..181] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [182..533] | 76.30103 | Crystal structure of MYO-inositol-1-phosphate synthase-related protein (TM1419) from Thermotoga maritima at 1.70 A resolution |
Functions predicted (by domain):
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)