













| General Information: |
|
| Name(s) found: |
MET13 /
YGL125W
[SGD]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 600 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA mitochondrion [IDA mitochondrial large ribosomal subunit [IDA |
| Biological Process: |
sulfur amino acid biosynthetic process
[TAS translation [IDA methionine metabolic process [IMP |
| Molecular Function: |
structural constituent of ribosome
[IDA methylenetetrahydrofolate reductase (NADPH) activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKITEKLEQH RQTSGKPTYS FEYFVPKTTQ GVQNLYDRMD RMYEASLPQF IDITWNAGGG 60
61 RLSHLSTDLV ATAQSVLGLE TCMHLTCTNM PISMIDDALE NAYHSGCQNI LALRGDPPRD 120
121 AENWTPVEGG FQYAKDLIKY IKSKYGDHFA IGVAGYPECH PELPNKDVKL DLEYLKQKID 180
181 AGGDFIITQM FYDVDNFINW CSQVRAAGMD VPIIPGIMPI TTYAAFLRRA QWGQISIPQH 240
241 FSSRLDPIKD DDELVRDIGT NLIVEMCQKL LDSGYVSHLH IYTMNLEKAP LMILERLNIL 300
301 PTESEFNAHP LAVLPWRKSL NPKRKNEEVR PIFWKRRPYS YVARTSQWAV DEFPNGRFGD 360
361 SSSPAFGDLD LCGSDLIRQS ANKCLELWST PTSINDVAFL VINYLNGNLK CLPWSDIPIN 420
421 DEINPIKAHL IELNQHSIIT INSQPQVNGI RSNDKIHGWG PKDGYVYQKQ YLEFMLPKTK 480
481 LPKLIDTLKN NEFLTYFAID SQGDLLSNHP DNSKSNAVTW GIFPGREILQ PTIVEKISFL 540
541 AWKEEFYHIL NEWKLNMNKY DKPHSAQFIQ SLIDDYCLVN IVDNDYISPD DQIHSILLSL 600
601 |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
MET12 MET13 |
| View Details | Gavin AC, et al. (2006) |

|
MET12 MET13 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
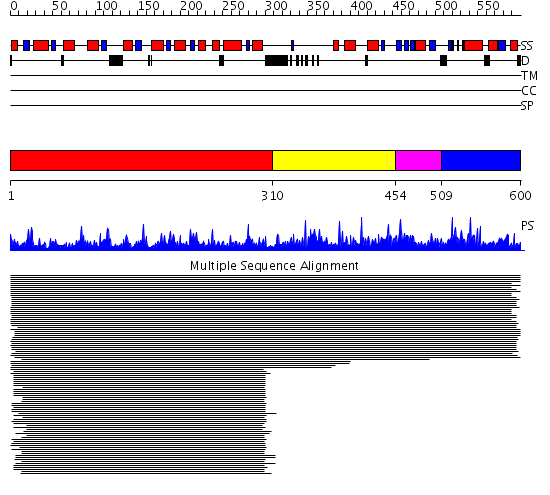
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..309] | 94.39794 | 5,10-Methylenetetrahydrofolate Reductase from Thermus thermophilus HB8 | |
| 2 | View Details | [310..453] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [454..508] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [509..600] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)