













| General Information: |
|
| Name(s) found: |
ATF1 /
YOR377W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 525 amino acids |
Gene Ontology: |
|
| Cellular Component: |
lipid particle
[IDA endomembrane system [TAS |
| Biological Process: |
acetate derivative biosynthetic process
[TAS fermentation [TAS fatty acid metabolic process [TAS |
| Molecular Function: |
alcohol O-acetyltransferase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNEIDEKNQA PVQQECLKEM IQNGHARRMG SVEDLYVALN RQNLYRNFCT YGELSDYCTR 60
61 DQLTLALREI CLKNPTLLHI VLPTRWPNHE NYYRSSEYYS RPHPVHDYIS VLQELKLSGV 120
121 VLNEQPEYSA VMKQILEEFK NSKGSYTAKI FKLTTTLTIP YFGPTGPSWR LICLPEEHTE 180
181 KWKKFIFVSN HCMSDGRSSI HFFHDLRDEL NNIKTPPKKL DYIFKYEEDY QLLRKLPEPI 240
241 EKVIDFRPPY LFIPKSLLSG FIYNHLRFSS KGVCMRMDDV EKTDDVVTEI INISPTEFQA 300
301 IKANIKSNIQ GKCTITPFLH VCWFVSLHKW GKFFKPLNFE WLTDIFIPAD CRSQLPDDDE 360
361 MRQMYRYGAN VGFIDFTPWI SEFDMNDNKE NFWPLIEHYH EVISEALRNK KHLHGLGFNI 420
421 QGFVQKYVNI DKVMCDRAIG KRRGGTLLSN VGLFNQLEEP DAKYSICDLA FGQFQGSWHQ 480
481 AFSLGVCSTN VKGMNIVVAS TKNVVGSQES LEELCSIYKA LLLGP |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |
|
ATF1 EHT1 SLC1 SUR4 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
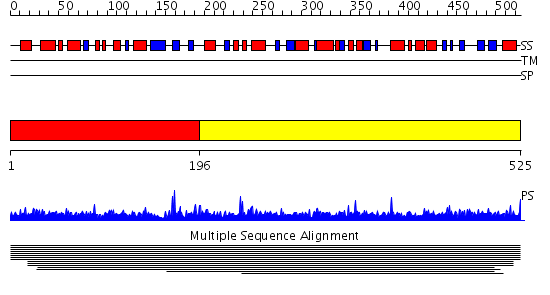
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..195] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [196..525] | 1.007918 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)