













| General Information: |
|
| Name(s) found: |
ZWF1 /
YNL241C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 505 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
pentose-phosphate shunt, oxidative branch
[ISS response to hydrogen peroxide [IMP |
| Molecular Function: |
glucose-6-phosphate dehydrogenase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEGPVKFEK NTVISVFGAS GDLAKKKTFP ALFGLFREGY LDPSTKIFGY ARSKLSMEED 60
61 LKSRVLPHLK KPHGEADDSK VEQFFKMVSY ISGNYDTDEG FDELRTQIEK FEKSANVDVP 120
121 HRLFYLALPP SVFLTVAKQI KSRVYAENGI TRVIVEKPFG HDLASARELQ KNLGPLFKEE 180
181 ELYRIDHYLG KELVKNLLVL RFGNQFLNAS WNRDNIQSVQ ISFKERFGTE GRGGYFDSIG 240
241 IIRDVMQNHL LQIMTLLTME RPVSFDPESI RDEKVKVLKA VAPIDTDDVL LGQYGKSEDG 300
301 SKPAYVDDDT VDKDSKCVTF AAMTFNIENE RWEGVPIMMR AGKALNESKV EIRLQYKAVA 360
361 SGVFKDIPNN ELVIRVQPDA AVYLKFNAKT PGLSNATQVT DLNLTYASRY QDFWIPEAYE 420
421 VLIRDALLGD HSNFVRDDEL DISWGIFTPL LKHIERPDGP TPEIYPYGSR GPKGLKEYMQ 480
481 KHKYVMPEKH PYAWPVTKPE DTKDN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
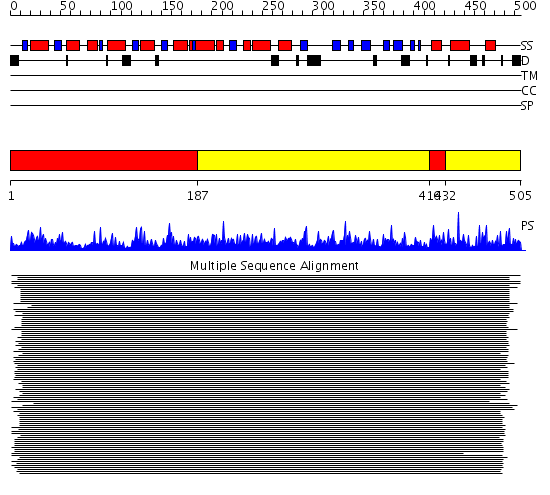
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..186] [416..431] |
2270.0 | Glucose 6-phosphate dehydrogenase, N-terminal domain; Glucose 6-phosphate dehydrogenase | |
| 2 | View Details | [187..415] [432..505] |
2270.0 | Glucose 6-phosphate dehydrogenase, N-terminal domain; Glucose 6-phosphate dehydrogenase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)