













| General Information: |
|
| Name(s) found: |
LEU4 /
YNL104C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 619 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA mitochondrion [IDA |
| Biological Process: |
leucine biosynthetic process
[IMP]
|
| Molecular Function: |
2-isopropylmalate synthase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVKESIIALA EHAASRASRV IPPVKLAYKN MLKDPSSKYK PFNAPKLSNR KWPDNRITRA 60
61 PRWLSTDLRD GNQSLPDPMS VEQKKEYFHK LVNIGFKEIE VSFPSASQTD FDFTRYAVEN 120
121 APDDVSIQCL VQSREHLIKR TVEALTGAKK ATIHTYLATS DMFREIVFNM SREEAISKAV 180
181 EATKLVRKLT KDDPSQQATR WSYEFSPECF SDTPGEFAVE ICEAVKKAWE PTEENPIIFN 240
241 LPATVEVASP NVYADQIEYF ATHITEREKV CISTHCHNDR GCGVAATELG MLAGADRVEG 300
301 CLFGNGERTG NVDLVTVAMN MYTQGVSPNL DFSDLTSVLD VVERCNKIPV SQRAPYGGDL 360
361 VVCAFSGSHQ DAIKKGFNLQ NKKRAQGETQ WRIPYLPLDP KDIGRDYEAV IRVNSQSGKG 420
421 GAAWVILRSL GLDLPRNMQI EFSSAVQDHA DSLGRELKSD EISKLFKEAY NYNDEQYQAI 480
481 SLVNYNVEKF GTERRVFTGQ VKVGDQIVDI EGTGNGPISS LVDALSNLLN VRFAVANYTE 540
541 HSLGSGSSTQ AASYIHLSYR RNADNEKAYK WGVGVSEDVG DSSVRAIFAT INNIIHSGDV 600
601 SIPSLAEVEG KNAAASGSA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
LEU4 LEU9 |
| View Details | Krogan NJ, et al. (2006) |
|
LEU4 LEU9 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
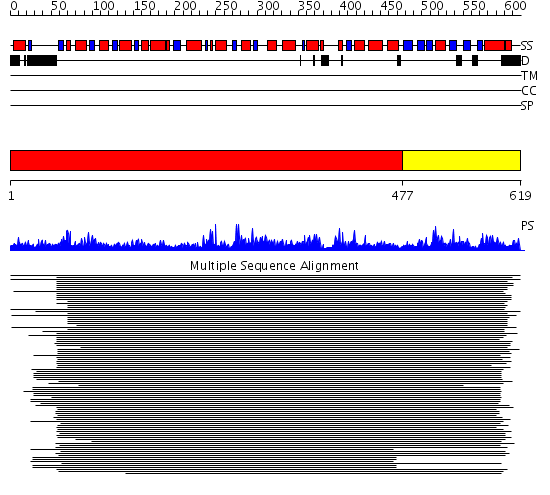
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..476] | 7.73 | Methylmalonyl-CoA mutase alpha subunit, domain 1; Methylmalonyl-CoA mutase alpha subunit, C-terminal domain | |
| 2 | View Details | [477..619] | 2.118979 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)