













| General Information: |
|
| Name(s) found: |
SCW10 /
YMR305C
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 389 amino acids |
Gene Ontology: |
|
| Cellular Component: |
fungal-type cell wall
[IDA cytoplasm [IDA endoplasmic reticulum [IDA |
| Biological Process: |
conjugation with cellular fusion
[IGI |
| Molecular Function: |
glucosidase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRFSNFLTVS ALLTGALGAP AVRHKHEKRD VVTATVHAQV TVVVSGNSGE TIVPVNENAV 60
61 VATTSSTAVA SQATTSTLEP TTSANVVTSQ QQTSTLQSSE AASTVGSSTS SSPSSSSSTS 120
121 SSASSSASSS ISASGAKGIT YSPYNDDGSC KSTAQVASDL EQLTGFDNIR LYGVDCSQVE 180
181 NVLQAKTSSQ KLFLGIYYVD KIQDAVDTIK SAVESYGSWD DITTVSVGNE LVNGGSATTT 240
241 QVGEYVSTAK SALTSAGYTG SVVSVDTFIA VINNPDLCNY SDYMAVNAHA YFDENTAAQD 300
301 AGPWVLEQIE RVYTACGGKK DVVITETGWP SKGDTYGEAV PSKANQEAAI SSIKSSCGSS 360
361 AYLFTAFNDL WKDDGQYGVE KYWGILSSD |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
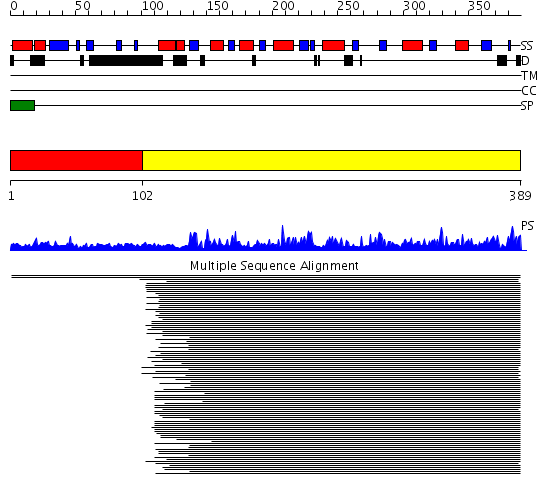
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..101] | 1.004993 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [102..389] | 102.0 | Plant beta-glucanases |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|