













| General Information: |
|
| Name(s) found: |
YML082W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 649 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
sulfur metabolic process
[ISS |
| Molecular Function: |
carbon-sulfur lyase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVSAQVATEL GQPIPLDTQH AVSVCFPTWK SVISYVEKDP KVLGCLKSGY PRFWIHPSIQ 60
61 KLRDILIEKY AKENETCFCF PSYRVAKRCR EYVRRKCAHR NGKVRILQLA TAKPINEEQK 120
121 TWKRECKIAV VFVDGAYENI LKQYWQYTGE IISSRLAEYV LHELFMVEKK SSPAEEKEYI 180
181 EMRYGRNLNF AFADRAKELI KKRIATKVID KDEHDEEENY HFLAGNQDEQ DFQDTFLDSS 240
241 LNEANHGEDH DGGISGEVDS QEEPHNGLVS TIPPEPIEMS TIEEEQSVEE DAGRCALRVC 300
301 PERDVFLFPS GMASIFTAHR LLLQWDSLRL NRSRNGSDVT SSPPNKKTVI FGFPYADTLH 360
361 VLQEFNETYF LGEGDESSMK ELTKILHSGE QILAVFIETP SNPLLKMGNL LELKRLSELF 420
421 GFFIIIDETV GGIVNIDGLP FADIVCSSLT KTFSGDSNVI GGSMVLNPQS RVYEFASRFM 480
481 QLEDEYEDLL WCEDAIYLER NSRDFIARTI RINYSTEYLL DKILKPHVGE NKLFKKIYYP 540
541 NLTSKETLTN YDMVRCKKEG GYGGLFSLTF HDEDHAAAFY DNLKLNKGPS LGTNFTLAFP 600
601 YTLMTYYHEL DMAEKFGVER NLLRISVGLE SQSILGKIFQ EAIDKTVEI |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
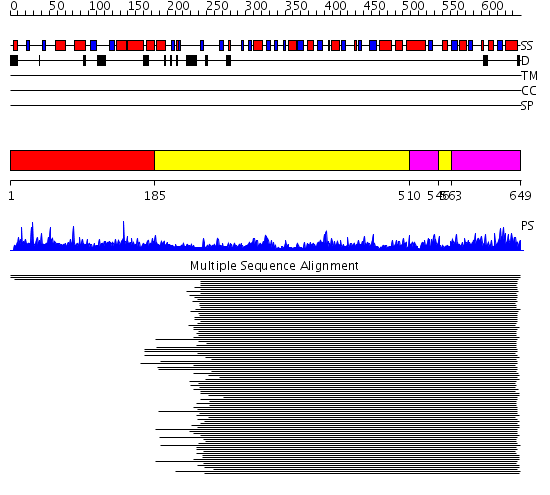
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..184] | 1.093935 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [185..509] [546..562] |
179.218487 | Cystathionine gamma-synthase, CGS | |
| 3 | View Details | [510..545] [563..649] |
179.218487 | Cystathionine gamma-synthase, CGS |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)