













| General Information: |
|
| Name(s) found: |
YLR455W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 304 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
response to drug
[IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTKDIRTGDL VLCKVGSFPP WPAVVFPQRL LRNDVYRKRK SNCVAVCFFN DPTYYWEQPS 60
61 RLKELDQDSI HNFILEHSKN ANQRELVNAY KEAKNFDDFN VFLQEKFEEE NRLSDLKAFE 120
121 KSEGSKIVAG EDPFVGRTKV VNKRKKNSIS IKEDPEDNQK SNEEESKPNI KPSKKKRPTA 180
181 NSGGKSNSGN KKKVKLDYSR RVEISQLFRR RIQRNLIQRE TPPTEHEIKE THELLNRIYE 240
241 NSDTKRPFFD LKALRESKLH KLLKAIVNDP DLGEFHPLCK EILLSWADLI TELKKEKLQA 300
301 LPTP |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
SAS3 TOP2 YLR455W |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
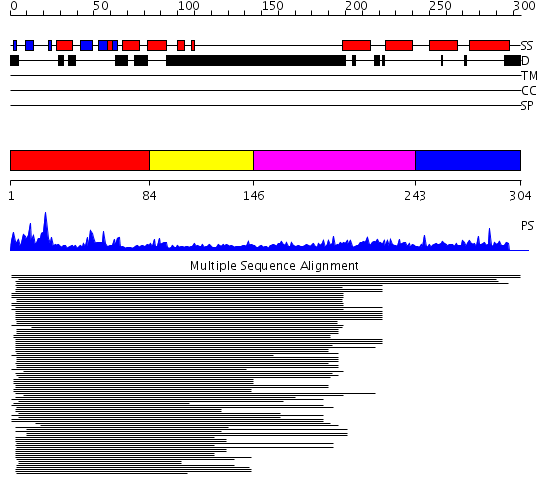
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..83] | 22.045757 | DNA methyltransferase DNMT3B | |
| 2 | View Details | [84..145] | 22.045757 | DNA methyltransferase DNMT3B | |
| 3 | View Details | [146..242] | 1.038994 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [243..304] | N/A | Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)