













| General Information: |
|
| Name(s) found: |
CTS1 /
YLR286C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 562 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear envelope
[IDA fungal-type cell wall [TAS endoplasmic reticulum [IDA extracellular region [IDA |
| Biological Process: |
cytokinesis, completion of separation
[IMP chitin catabolic process [IC |
| Molecular Function: |
endochitinase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLLYIILLF TQFLLLPTDA FDRSANTNIA VYWGQNSAGT QESLATYCES SDADIFLLSF 60
61 LNQFPTLGLN FANACSDTFS DGLLHCTQIA EDIETCQSLG KKVLLSLGGA SGSYLFSDDS 120
121 QAETFAQTLW DTFGEGTGAS ERPFDSAVVD GFDFDIENNN EVGYSALATK LRTLFAEGTK 180
181 QYYLSAAPQC PYPDASVGDL LENADIDFAF IQFYNNYCSV SGQFNWDTWL TYAQTVSPNK 240
241 NIKLFLGLPG SASAAGSGYI SDTSLLESTI ADIASSSSFG GIALWDASQA FSNELNGEPY 300
301 VEILKNLLTS ASQTATTTVA TSKTSAASTS SASTSSASTS QKKTTQSTTS TQSKSKVTLS 360
361 PTASSAIKTS ITQTTKTLTS STKTKSSLGT TTTESTLNSV AITSMKTTLS SQITSAALVT 420
421 PQTTTTSIVS SAPIQTAITS TLSPATKSSS VVSLQTATTS TLSPTTTSTS SGSTSSGSTS 480
481 SDSTARTLAK ELNAQYAAGK LNGKSTCTEG EIACSADGKF AVCDHSAWVY MECASGTTCY 540
541 AYDSGDSVYT QCNFSYLESN YF |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |
|
CTS1 DSE4 PGU1 SUC2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #06 Alpha Factor Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
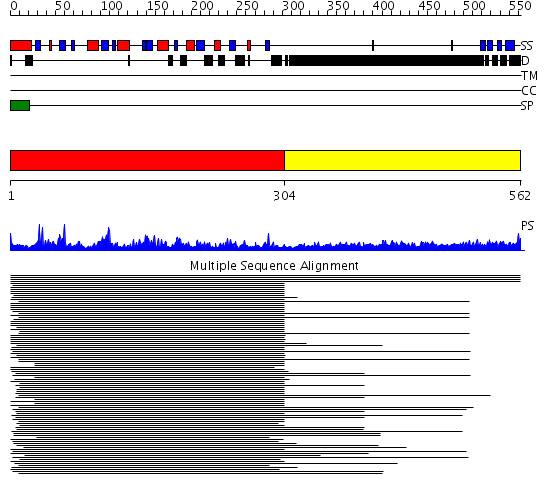
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..303] | 459.40824 | Hevamine A (chitinase/lysozyme) | |
| 2 | View Details | [304..562] | 14.75 | Sec24 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|