













| General Information: |
|
| Name(s) found: |
HUL4 /
YJR036C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 892 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
protein polyubiquitination
[TAS protein monoubiquitination [TAS |
| Molecular Function: |
ubiquitin-protein ligase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVSLFDKLNA KKDGRDGSVS KELLSHSVAH TKNRLPKSGR RTSERSLAAS VKDGSCSNSK 60
61 SNKRNSSASV SGEEDKSCLI SLNCLCCGVP LRFPASITKF RCSACQVTVI VKEPEINSNL 120
121 ESSTHISCTL EGLQMVVRRC HDDLQRLKKT GILDKERKGL IFQPVITYLL DRFHDVSILN 180
181 RSFLVHDGGK NIKMLNYEVL QRFYSILSNL PTRKPYYSML CCCNDLLKRI TINKGENLQI 240
241 LQYRWLLIIL NIPTIRTCLI RDRKSKNVFE TQQIRAVSYE LAKRCIGYLS NLSTKTSQQL 300
301 IQSLRRTPTD NFSYQVEILN LYINFQFSRL LSNELSNRTA KNNVKPEDEM RSRLRRHHTT 360
361 GHEFLSTRPI SAQSNDKQGS GFTHPVNNKM KFKFFQYEED WHIHSAAKLT FIYYVANTRR 420
421 NGRGALSIQS FYNITLDFID YKQDFDHWRG VAQKTKMNQL IEEWGNSTTK KCFSFCKYPF 480
481 ILSLGIKISI MEYEIRRIME HEAEQAFLIS LDKGKSVDVY FKIKVRRDVI SHDSLRCIKE 540
541 HQGDLLKSLR IEFVNEPGID AGGLRKEWFF LLTKSLFNPM NGLFIYIKES SRSWFAIDPP 600
601 NFDKSKGKNS QLELYYLFGV VMGLAIFNST ILDLQFPKAL YKKLCSEPLS FEDYSELFPE 660
661 TSRNLIKMLN YTEDNFEDVF SLTFETTYRN NNWILNDSKS SKEYVTVELC ENGRNVPITQ 720
721 SNKHEFVMKW VEFYLEKSIE PQYNKFVSGF KRVFAECNSI KLFNSEELER LVCGDEEQTK 780
781 FDFKSLRSVT KYVGGFSDDS RAVCWFWEII ESWDYPLQKK LLQFVTASDR IPATGISTIP 840
841 FKISLLGSHD SDDLPLAHTC FNEICLWNYS SKKKLELKLL WAINESEGYG FR |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
HUL4 MCK1 PRD1 YGR012W |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
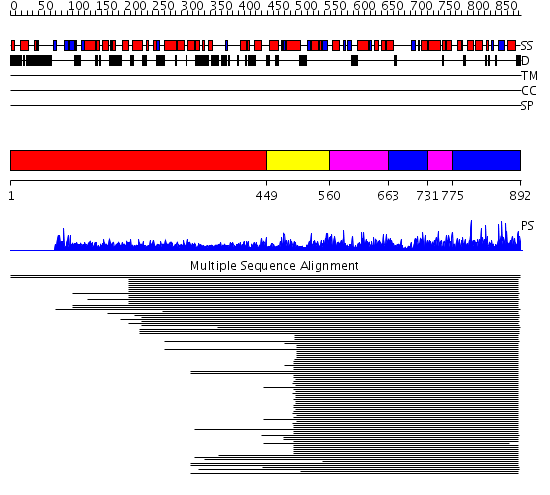
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..448] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [449..559] | 710.0103 | Ubiquitin-protein ligase E3a, Hect catalytic domain (E6ap) | |
| 3 | View Details | [560..662] [731..774] |
710.0103 | Ubiquitin-protein ligase E3a, Hect catalytic domain (E6ap) | |
| 4 | View Details | [663..730] [775..892] |
710.0103 | Ubiquitin-protein ligase E3a, Hect catalytic domain (E6ap) | |
| 5 | View Details | [509..892] | 122.0 | Ubiquitin-protein ligase E3a, Hect catalytic domain (E6ap) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.72 |
Source: Reynolds et al. (2008)