













| General Information: |
|
| Name(s) found: |
MAD3 /
YJL013C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 515 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
mitotic cell cycle spindle assembly checkpoint
[IGI |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKAYAKKRIS YMPSSPSQNV INFEEIETQK ENILPLKEGR SAAALSKAIH QPLVEINQVK 60
61 SSFEQRLIDE LPALSDPITL YLEYIKWLNN AYPQGGNSKQ SGMLTLLERC LSHLKDLERY 120
121 RNDVRFLKIW FWYIELFTRN SFMESRDIFM YMLRNGIGSE LASFYEEFTN LLIQKEKFQY 180
181 AVKILQLGIK NKARPNKVLE DRLNHLLREL GENNIQLGNE ISMDSLESTV LGKTRSEFVN 240
241 RLELANQNGT SSDVNLTKNN VFVDGEESDV ELFETPNRGV YRDGWENFDL KAERNKENNL 300
301 RISLLEANTN LGELKQHEML SQKKRPYDEK LPIFRDSIGR SDPVYQMINT KDQKPEKIDC 360
361 NFKLIYCEDE ESKGGRLEFS LEEVLAISRN VYKRVRTNRK HPREANLGQE ESANQKEAEA 420
421 QSKRPKISRK ALVSKSLTPS NQGRMFSGEE YINCPMTPKG RSTETSDIIS AVKPRQLTPI 480
481 LEMRESNSFS QSKNSEIISD DDKSSSSFIS YPPQR |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Ho Y, et al. (2002) |
|
CCT2 CCT3 CCT5 CDC20 MAD3 MDH1 MKK2 TCP1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
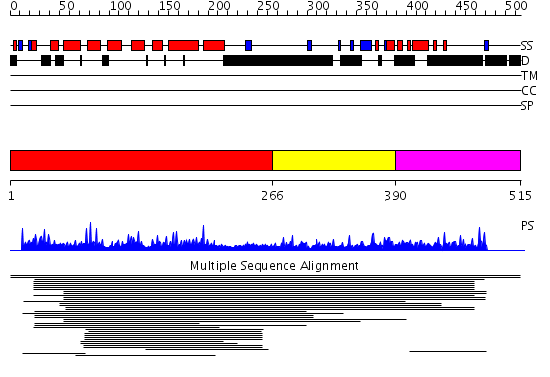
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..265] | 7.6 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 2 | View Details | [266..389] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [390..515] | 1.009976 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)