













| General Information: |
|
| Name(s) found: |
DJP1 /
YIR004W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 432 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA |
| Biological Process: |
protein import into peroxisome matrix
[IMP |
| Molecular Function: |
chaperone binding
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVVDTEYYDL LGVSTTASSI EIKKAYRKKS IQEHPDKNPN DPTATERFQA ISEAYQVLGD 60
61 DDLRAKYDKY GRKEAIPQGG FEDAAEQFSV IFGGDAFASY IGELMLLKNL QKTEELNAED 120
121 EAEKEKENVE TMEESPADGK TNGTTNAVDA ALGNTNEKDD KNKARTTSGN LTVHDGNKKN 180
181 EQVGAEAKKK KTKLEQFEEE QEVEKQKRVD QLSKTLIERL SILTESVYDD ACKDSFKKKF 240
241 EEEANLLKME SFGLDILHTI GDVYYEKAEI FLASQNLFGM GGIFHSMKAK GGVFMDTLRT 300
301 VSAAIDAQNT MKELEKMKEA STNNEPLFDK DGNEQIKPTT EELAQQEQLL MGKVLSAAWH 360
361 GSKYEITSTL RGVCKKVLED DSVSKKTLIR RAEAMKLLGE VFKKTFRTKV EQEEAQIFEE 420
421 LVAEATKKKR HT |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | De Craene, J., et al. (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
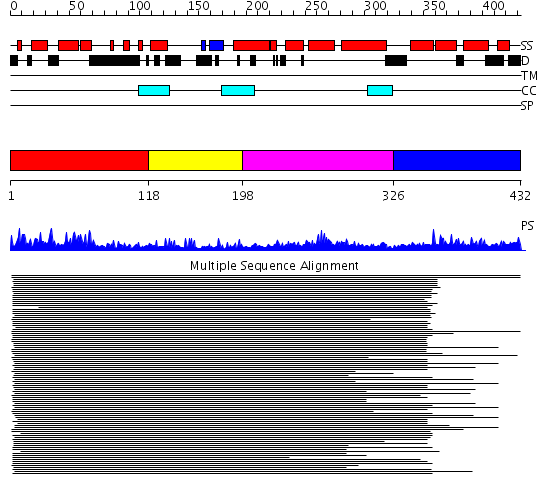
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..117] | 143.70927 | DnaJ chaperone, N-terminal (J) domain | |
| 2 | View Details | [118..197] | 17.69897 | Cysteine-rich domain of the chaperone protein DnaJ. | |
| 3 | View Details | [198..325] | 15.045757 | Heat shock protein 40 Sis1 | |
| 4 | View Details | [326..432] | 1.150976 | View MSA. Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)