













| General Information: |
|
| Name(s) found: |
ASG1 /
YIL130W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 964 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPEQAQQGEQ SVKRRRVTRA CDECRKKKVK CDGQQPCIHC TVYSYECTYK KPTKRTQNSG 60
61 NSGVLTLGNV TTGPSSSTVV AAAASNPNKL LSNIKTERAI LPGASTIPAS NNPSKPRKYK 120
121 TKSTRLQSKI DRYKQIFDEV FPQLPDIDNL DIPVFLQIFH NFKRDSQSFL DDTVKEYTLI 180
181 VNDSSSPIQP VLSSNSKNST PDEFLPNMKS DSNSASSNRE QDSVDTYSNI PVGREIKIIL 240
241 PPKAIALQFV KSTWEHCCVL LRFYHRPSFI RQLDELYETD PNNYTSKQMQ FLPLCYAAIA 300
301 VGALFSKSIV SNDSSREKFL QDEGYKYFIA ARKLIDITNA RDLNSIQAIL MLIIFLQCSA 360
361 RLSTCYTYIG VAMRSALRAG FHRKLSPNSG FSPIEIEMRK RLFYTIYKLD VYINAMLGLP 420
421 RSISPDDFDQ TLPLDLSDEN ITEVAYLPEN QHSVLSSTGI SNEHTKLFLI LNEIISELYP 480
481 IKKTSNIISH ETVTSLELKL RNWLDSLPKE LIPNAENIDP EYERANRLLH LSFLHVQIIL 540
541 YRPFIHYLSR NMNAENVDPL CYRRARNSIA VARTVIKLAK EMVSNNLLTG SYWYACYTIF 600
601 YSVAGLLFYI HEAQLPDKDS AREYYDILKD AETGRSVLIQ LKDSSMAASR TYNLLNQIFE 660
661 KLNSKTIQLT ALHSSPSNES AFLVTNNSSA LKPHLGDSLQ PPVFFSSQDT KNSFSLAKSE 720
721 ESTNDYAMAN YLNNTPISEN PLNEAQQQDQ VSQGTTNMSN ERDPNNFLSI DIRLDNNGQS 780
781 NILDATDDVF IRNDGDIPTN SAFDFSSSKS NASNNSNPDT INNNYNNVSG KNNNNNNITN 840
841 NSNNNHNNNN NDNNNNNNNN NNNNNNNNNS GNSSNNNNNN NNNKNNNDFG IKIDNNSPSY 900
901 EGFPQLQIPL SQDNLNIEDK EEMSPNIEIK NEQNMTDSND ILGVFDQLDA QLFGKYLPLN 960
961 YPSE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
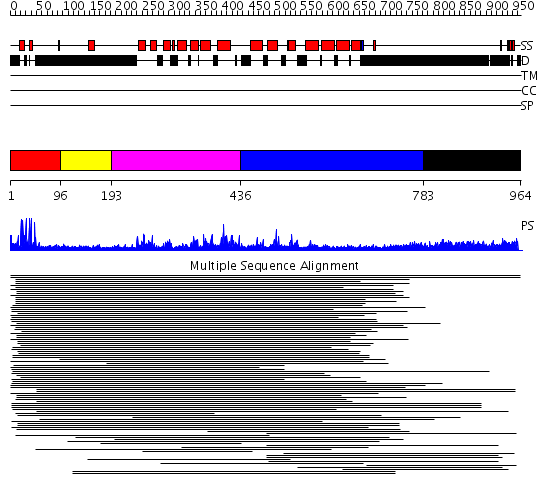
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..95] | 44.68867 | Gal4; CD2-Gal4 | |
| 2 | View Details | [96..192] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [193..435] | 3.107996 | View MSA. Confident ab initio structure predictions are available. | |
| 4 | View Details | [436..782] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [783..964] | 18.103997 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [697..964] | 2.17 | 30S subunit |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. | ||||||
| 5 | No functions predicted. | ||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.67 |
Source: Reynolds et al. (2008)