













| General Information: |
|
| Name(s) found: |
YIL092W
[SGD]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 633 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVQMRSKNMA YESGTNNYSD TIANGNTLPP RSKKGHSGRR KRSETLPIAC NNFCVTRQID 60
61 DDEQAFKMLD KVSHLKKFSA EDGDDNNIFV QWADDITDIL FGLCCTGTFL KLLISSALSG 120
121 RAKTWFDSTT EGIDDHVIKA YSFEKFLALL SEEFDGARSL RREIFTELLT LSIDSEKSLE 180
181 AFAHKSGRLT PYYLSSGAAL DLFLTKLEPQ LQKQLENCAF PMTLNLALLI TACEFAKRAS 240
241 NHKKYRYKNT RDSDICTPKS KNTAIVSKLS NTKTISKNKV IEKSDKKNYF DKNSQHIPDP 300
301 KRRKQNEPGM RLFLVMDEEK NILTSRNVSA NAYTSKNGHT NLSDLHTNLK NSKSQQCAVE 360
361 PISILNSGSL VTGTINIDLI NDEVLGTKEE TTTYDERMDG NSRSLNERCC AVKKNSLQPI 420
421 TSNIFQKNAE IQGTKIGSVL DSGISNSFSS TEYMFPPTSS ATVSNPVKKN EISKSSQVKD 480
481 IAQFNPFMTN EKEKKLNPSE SFKSPGVSME INRLSRIAGL RNIPGNIYED SKMLNLKTRK 540
541 CYPLHNFAVR TRSAHFNDRP SNYISPHETI NATLRSPASF DSIQCITRSK RVDAETNKAT 600
601 GSAKSENIET KSRKFPEVIN PFLVNTTNKK ESD |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
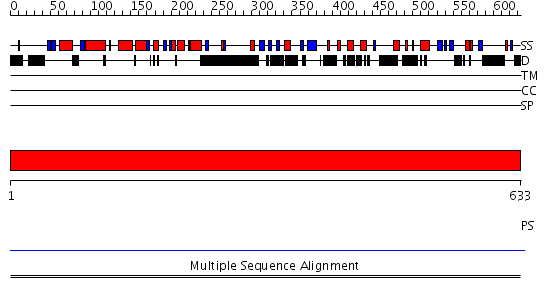
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..633] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [269..458] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [459..633] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)