













| General Information: |
|
| Name(s) found: |
TRM5 /
YHR070W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 499 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
ribosome biogenesis
[RCA tRNA methylation [IGI |
| Molecular Function: |
tRNA (guanine) methyltransferase activity
[IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKIALPVFQK FNRLISSCKM SGVFPYNPPV NRQMRELDRS FFITKIPMCA VKFPEPKNIS 60
61 VFSKNFKNCI LRVPRIPHVV KLNSSKPKDE LTSVQNKKLK TADGNNTPVT KGVLLHESIH 120
121 SVEDAYGKLP EDALAFLKEN SAEIVPHEYV LDYDFWKAEE ILRAVLPEQF LEEVPTGFTI 180
181 TGHIAHLNLR TEFKPFDSLI GQVILDKNNK IECVVDKVSS IATQFRTFPM KVIAGKSDSL 240
241 VVEQKESNCT FKFDFSKVYW NSRLHTEHER LVKQYFQPGQ VVCDVFAGVG PFAVPAGKKD 300
301 VIVLANDLNP ESYKYLKENI ALNKVAKTVK SFNMDGADFI RQSPQLLQQW IQDEEGGKIT 360
361 IPLPLKKRHR SQQHNDQQPP QPRTKELIIP SHISHYVMNL PDSAISFLGN FRGIFAAHTK 420
421 GATDTIQMPW VHVHCFEKYP PGDQVTEDEL HARVHARIIA ALKVTADDLP LNAVSLHLVR 480
481 KVAPTKPMYC ASFQLPANV |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
ATG18 TRM5 |
| View Details | Qiu et al. (2008) |

|
TRM10 TRM5 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Drees BL, et al. (2001) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
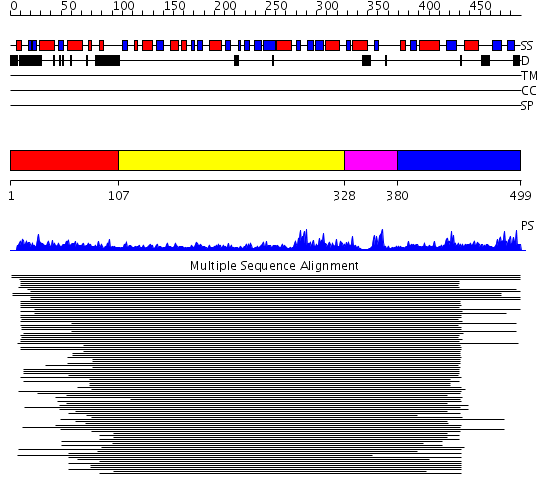
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..106] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [107..327] | 6.69897 | Methyltransferase mboII | |
| 3 | View Details | [328..379] | 14.221849 | Hypothetical protein MJ0882 | |
| 4 | View Details | [380..499] | N/A | Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)