













| General Information: |
|
| Name(s) found: |
UGA1 /
YGR019W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 471 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IC |
| Biological Process: |
nitrogen utilization
[TAS |
| Molecular Function: |
4-aminobutyrate transaminase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSICEQYYPE EPTKPTVKTE SIPGPESQKQ LKELGEVFDT RPAYFLADYE KSLGNYITDV 60
61 DGNTYLDLYA QISSIALGYN NPALIKAAQS PEMIRALVDR PALGNFPSKD LDKILKQILK 120
121 SAPKGQDHVW SGLSGADANE LAFKAAFIYY RAKQRGYDAD FSEKENLSVM DNDAPGAPHL 180
181 AVLSFKRAFH GRLFASGSTT CSKPIHKLDF PAFHWPHAEY PSYQYPLDEN SDANRKEDDH 240
241 CLAIVEELIK TWSIPVAALI IEPIQSEGGD NHASKYFLQK LRDITLKYNV VYIIDEVQTG 300
301 VGATGKLWCH EYADIQPPVD LVTFSKKFQS AGYFFHDPKF IPNKPYRQFN TWCGEPARMI 360
361 IAGAIGQEIS DKKLTEQCSR VGDYLFKKLE GLQKKYPENF QNLRGKGRGT FIAWDLPTGE 420
421 KRDLLLKKLK LNGCNVGGCA VHAVRLRPSL TFEEKHADIF IEALAKSVNE L |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | hx21: both fusion and trans-SNARE complex formation take place. Investigating Vam3p, Nyv1p and their partners in trans-SNARE complex | Hao Xu, et al. (2010) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
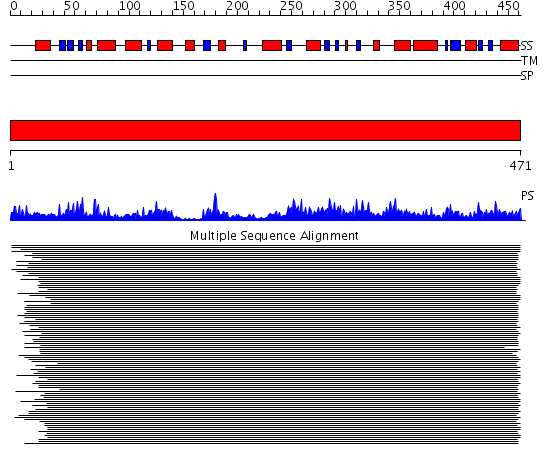
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..471] | 1210.0 | 4-aminobutyrate aminotransferase, GABA-aminotransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)