













| General Information: |
|
| Name(s) found: |
PTR3 /
YFR029W
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 678 amino acids |
Gene Ontology: |
|
| Cellular Component: |
extrinsic to plasma membrane
[IDA |
| Biological Process: |
response to amino acid stimulus
[IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHSHRQKWGR QTDIARVLDD IEHDLYLPQR LSLDGATGTD ESHVQYGIVK DCSVLTCGCC 60
61 ISESLFNDLC RETSNKQTAC PICQRENVRL LSAIKPLRDL ARQIDFFRST TGQGESESDE 120
121 LPAIVKTSPS SSSLSLTPSR SSSTAGLEAD NKTLSDPTVK EKSSLLELFH IVASKMHNAN 180
181 TEVGSDHPLT TGTTRDQEEH TTKENYSSSL LEPNYDDHAN WKILDNASNT RTVPIDNNFS 240
241 LMSTDVTIPS TANYQTNSAH DLDEEKEYFF ANCFPMYRKK FQFNTHPKFL GTKSKLFINQ 300
301 SISPDCTKFA LITEHKWEIY SINPKDNSPQ LVSCGKSSGE YGPNFNQLTE QSSSSLSTTS 360
361 QASKKKKKNW SQRFCKLSND FLIISGSQNI LNVHDIHQNG KLIYTYVSRF PIRCIDIDPR 420
421 SQIIAYGITG KDRHTGAEQA LVVIQQITRN KVTLEPEFPP PITITLPYRD PINTIQLSHD 480
481 AKYLTCSTAL ESRFLIISLQ KINEPRLIMK SVRSIDTSLE SEGITDTKLF PGNPNLMCIT 540
541 STAFNSSPLV INTKITQING VRTVAQPSML IRVDEIGCKI HKCEISPRND AIAFLDRNGS 600
601 VYIMCAPTMM DNNEKRRTIL VETVANAYRA YESATLRFNP EGNKLYILDR KGTFFVEDFA 660
661 YGLPQSREIT KCKQIFHK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |

|
PTR3 SSY1 SSY5 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #34 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
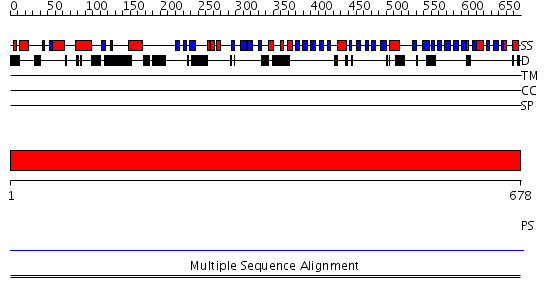
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..678] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [147..219] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [220..678] | 1.03 | N-terminal (heme c) domain of cytochrome cd1-nitrite reductase; C-terminal (heme d1) domain of cytochrome cd1-nitrite reductase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||
| 1 |
|
||||||||||||
| 2 | No functions predicted. | ||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)