













| General Information: |
|
| Name(s) found: |
SPS2 /
YDR522C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 502 amino acids |
Gene Ontology: |
|
| Cellular Component: |
fungal-type cell wall
[IDA |
| Biological Process: |
ascospore formation
[IEP ascospore wall assembly [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPIWKTQTFF TSISVIQIVN KETKVSTKKE KDSMLNQLNT ILRFLFLFLQ LIKSSAAVEP 60
61 NGGPNILDHN IMLVNTNATI PKKEQTDFEV ISPTKQTQVD EDCKKGLYHI ENAGNLIELQ 120
121 AKCWKVVGNI EISSNYSGSL IDLGLIREIE GDLIIKNNKH IFRIQGYNLE SLGKLELDSL 180
181 TSFVSLDFPA LKEVETVDWR VLPILSSVVI NGNIKKIKNI IISDTALTSI DYFNNVKKVD 240
241 IFNINNNRFL ENLFASLESV TKQLTVHSNA KELELDLSNL HTVENMTIKD VSEIKLAKLS 300
301 SVNSSLEFIE NQFSSLELPL LAKVQGTLGL IDNKNLKKLN FSNATDIQGG LMIANNTELA 360
361 KIDFFPKLRQ IGGAIYFEGS FDKIDLPELK LVKGSAYIKS SSEELNCEEF TSPKAGRSII 420
421 RGGKIECTSG MKSKMLNVDE EGNVLGKQET DNDNGKKEKG KNGAKSQGSS KKMENSAPKN 480
481 IFIDAFKMSV YAVFTVLFSI IF |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
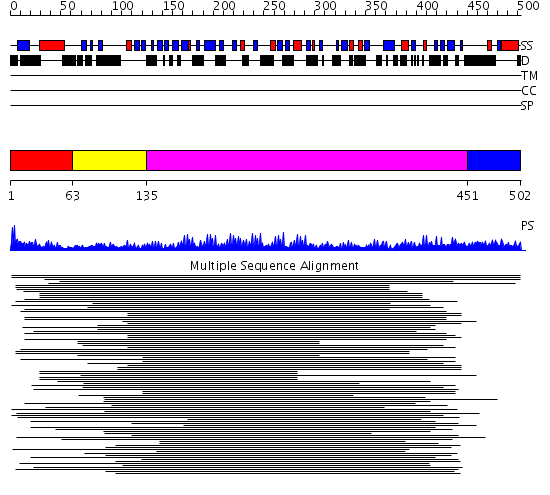
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..62] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [63..134] | 26.154902 | Internalin H | |
| 3 | View Details | [135..450] | 15.0 | Leucine rich effector protein YopM | |
| 4 | View Details | [451..502] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)