













| General Information: |
|
| Name(s) found: |
ASP1 /
YDR321W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 381 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[TAS]
intracellular [IDA |
| Biological Process: |
asparagine catabolic process
[IGI |
| Molecular Function: |
asparaginase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKSDSVEITT ICPDVENSQF VVQSNCPETI PEILKSQNAA VNGSGIACQQ RSLPRIKILG 60
61 TGGTIASKAI DSSQTAGYHV DLTIQDLLDA IPDISKVCDI EYEQLCNVDS KDINEDILYK 120
121 IYKGVSESLQ AFDGIVITHG TDTLSETAFF IESTIDAGDV PIVFVGSMRP STSVSADGPM 180
181 NLYQAICIAS NPKSRGRGVL VSLNDQISSG YYITKTNANS LDSFNVRQGY LGNFVNNEIH 240
241 YYYPPVKPQG CHKFKLRVDG KHFKLPEVCI LYAHQAFPPA IVNLVADKYD GIVLATMGAG 300
301 SLPEEVNETC MKLSLPIVYS KRSMDGMVPI ANVPKKGSKE DNLIASGYLS PEKSRILLQL 360
361 CLAGNYTLEE IKHVFTGVYG G |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | #08 Alpha Factor Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Hazbun TR, et al. (2003) | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Hazbun TR, et al. (2003) |
New Feature: Upload Your Own Microscopy Data
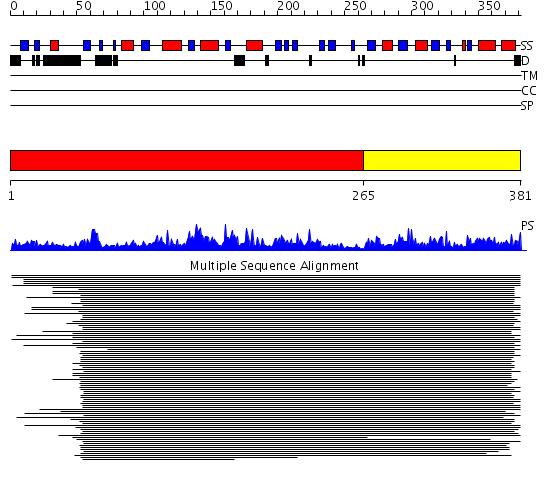
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..264] | 609.9897 | Asparaginase type II | |
| 2 | View Details | [265..381] | 609.9897 | Asparaginase type II |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 |
|
||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)