













| General Information: |
|
| Name(s) found: |
DPL1 /
YDR294C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 589 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum
[IDA |
| Biological Process: |
cellular response to starvation
[IMP calcium-mediated signaling [IMP sphingolipid metabolic process [IGI |
| Molecular Function: |
sphinganine-1-phosphate aldolase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGVSNKTVS INGWYGMPIH LLREEGDFAQ FMILTINELK IAIHGYLRNT PWYNMLKDYL 60
61 FVIFCYKLIS NFFYLLKVYG PVRLAVRTYE HSSRRLFRWL LDSPFLRGTV EKEVTKVKQS 120
121 IEDELIRSDS QLMNFPQLPS NGIPQDDVIE ELNKLNDLIP HTQWKEGKVS GAVYHGGDDL 180
181 IHLQTIAYEK YCVANQLHPD VFPAVRKMES EVVSMVLRMF NAPSDTGCGT TTSGGTESLL 240
241 LACLSAKMYA LHHRGITEPE IIAPVTAHAG FDKAAYYFGM KLRHVELDPT TYQVDLGKVK 300
301 KFINKNTILL VGSAPNFPHG IADDIEGLGK IAQKYKLPLH VDSCLGSFIV SFMEKAGYKN 360
361 LPLLDFRVPG VTSISCDTHK YGFAPKGSSV IMYRNSDLRM HQYYVNPAWT GGLYGSPTLA 420
421 GSRPGAIVVG CWATMVNMGE NGYIESCQEI VGAAMKFKKY IQENIPDLNI MGNPRYSVIS 480
481 FSSKTLNIHE LSDRLSKKGW HFNALQKPVA LHMAFTRLSA HVVDEICDIL RTTVQELKSE 540
541 SNSKPSPDGT SALYGVAGSV KTAGVADKLI VGFLDALYKL GPGEDTATK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | experimental2 - mih1-2 | McCusker D, et al (2007) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
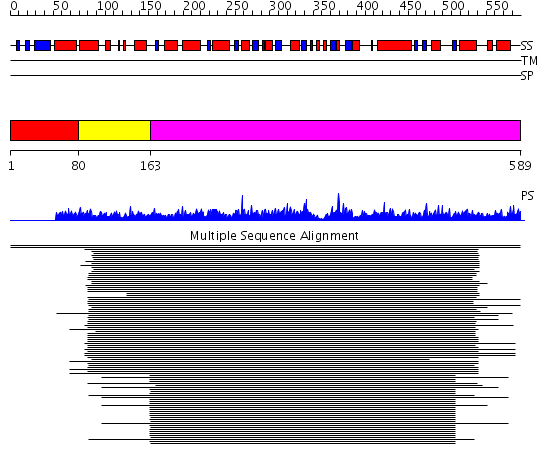
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..79] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [80..162] | 107.0 | DOPA decarboxylase | |
| 3 | View Details | [163..589] | 107.0 | DOPA decarboxylase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)