













| General Information: |
|
| Name(s) found: |
SAS4 /
YDR181C
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 481 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear chromatin
[IDA nuclear chromosome, telomeric region [IC |
| Biological Process: |
chromatin silencing at telomere
[IDA |
| Molecular Function: |
acetyltransferase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGIFQSIEEA NNTSERLLRS EIKNSHGEFE KFDFDTEEYE INPKRKLRLV SRINPNAGHL 60
61 RKSKSCFTVD EHVDETRCQK PVMKSPFMSN VDDEIKKKRE TITKMTLEIE HHELSQNIRK 120
121 PTDDLLPDST YQPYHKKMLK QENRMIQSDI VNGENEADRL SLISDRLGML NWEVTLQKVT 180
181 KINDPTDENE METKRYQTKE LIDSMLHKFE SMKKKSRNLA RRPASSDSLL KLVSGKDWPK 240
241 IYTRIDRTFI PDYASSSDEE EEKITVEEIR ERRLKKREQQ CGGSIIVLLS DHQSQKGMTR 300
301 FAIVAEPLRK PYLIKTSTKE RNSWKNKVPT NPKKFKKAPR ISTQIAVKRR REVIPLTMEV 360
361 EPEVIRDIRQ DTQKSMKLNV KAEEISVTET VKSKEMNALR NNAASISPTL SEKAPLGSIS 420
421 SCTASQISQR SSENVGAIIN NINPNLAIVP SCNEKTFVKT HNGMKTNSGI NILPVRKKKK 480
481 V |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
SAS2 SAS4 |
| View Details | Qiu et al. (2008) |
|
ARP4 PCC1 SAS3 SAS4 SAS5 YNG1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
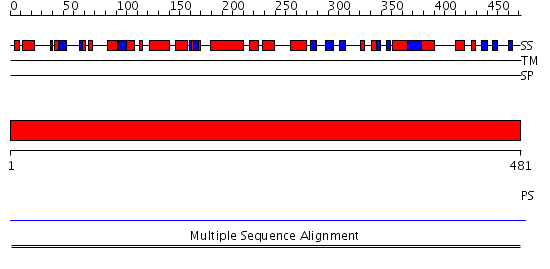
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..481] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)