













| General Information: |
|
| Name(s) found: |
PHO2 /
YDL106C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 559 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
transcription
[IGI histidine biosynthetic process [TAS purine base biosynthetic process [IDA cellular response to phosphate starvation [TAS |
| Molecular Function: |
transcription factor activity
[IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMEEFSYDHD FNTHFATDLD YLQHDQQQQQ QQQHDQQHNQ QQQPQPQPIQ TQNLEHDHDQ 60
61 HTNDMSASSN ASDSGPQRPK RTRAKGEALD VLKRKFEINP TPSLVERKKI SDLIGMPEKN 120
121 VRIWFQNRRA KLRKKQHGSN KDTIPSSQSR DIANDYDRGS TDNNLVTTTS TSSIFHDEDL 180
181 TFFDRIPLNS NNNYYFFDIC SITVGSWNRM KSGALQRRNF QSIKELRNLS PIKINNIMSN 240
241 ATDLMVLISK KNSEINYFFS AMANNTKILF RIFFPLSSVT NCSLTLETDD DIINSNNTSD 300
301 KNNSNTNNDD DNDDNSNEDN DNSSEDKRNA KDNFGELKLT VTRSPTFAVY FLNNAPDEDP 360
361 NLNNQWSICD DFSEGRQVND AFVGGSNIPH TLKGLQKSLR FMNSLILDYK SSNEILPTIN 420
421 TAIPTAAVPQ QNIAPPFLNT NSSATDSNPN TNLEDSLFFD HDLLSSSITN TNNGQGSNNG 480
481 RQASKDDTLN LLDTTVNSNN NHNANNEENH LAQEHLSNDA DIVANPNDHL LSLPTDSELP 540
541 NTPDFLKNTN ELTDEHRWI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
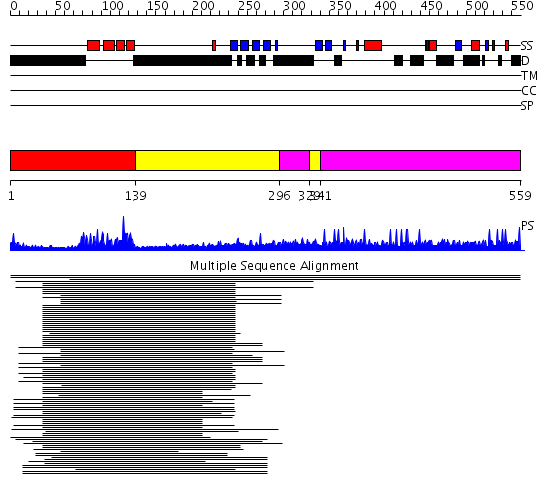
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..138] | 83.529817 | Paired protein | |
| 2 | View Details | [139..295] [329..340] |
7.23 | Adenovirus hexon | |
| 3 | View Details | [296..328] [341..559] |
7.23 | Adenovirus hexon | |
| 4 | View Details | [458..559] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)