













| General Information: |
|
| Name(s) found: |
DSF2 /
YBR007C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 736 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNQNLKNTSW ADRIGSDDQE RKANSSEVSQ SPPPNNSFES SMDSQFSYAH SNKSSISFES 60
61 IQTTERLLDK LDLSLEDELI LQEALLEEEN ASRNSQLSQT SGPTLCMPAS EFPSLRYRTN 120
121 PSPTYIQARD RSLIIDNLKE KDSTLRGKYS SGKVERHLPV KSRYSYIVEE DYDSETFSGM 180
181 KPQMNRNEKD YKYPNLENGN RSTNRPNPFN FEKYRIENTR LHHLYPTLIS DNNTSVDNNA 240
241 NSKNNRTTSN NINTSTKTDR ISEKQSCPNE FTTTQKSNCL YRNGSSTSTN TSFSEVGQLS 300
301 KPKTQSSFES ESSSFSKLKL TKSDTTTIKP SPKRSNSSTS TITKTNTMTN DISLPPTPPY 360
361 KAHKKKTSLN SLKKLFKSPR TRAKNKKDLE SEGSSPIRSA TNSLDFSGEN IQLPSTSSTI 420
421 NNSSPHLARY IFPPNPVFHF KTASTPQSST DKKKNSKARP NRTHLRTFSD FHTTEKDSKI 480
481 GELSALTEQS NKPYHPKVRR RTLSLDGMLP NNSTQCMDSF SHKKEGSNAT SKCGKLKFHP 540
541 EPYDNDESSH IGQAITMRHQ GKLEESAQRL KKACACGNKT AFLLYGLALR HGCGVDKNLK 600
601 LSLGYLMAAT DIKSFAAEVL DLDINPLNFA SMDDIPDIAP EPTAPALYEC GMAYLKGLGM 660
661 DHPDERKGLK FLEKAALLGH VDSMCLSGTI WSKTSNVKKR DLARAAAWFR IADKKGANLL 720
721 GSDWIYKEKY MKQGPK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
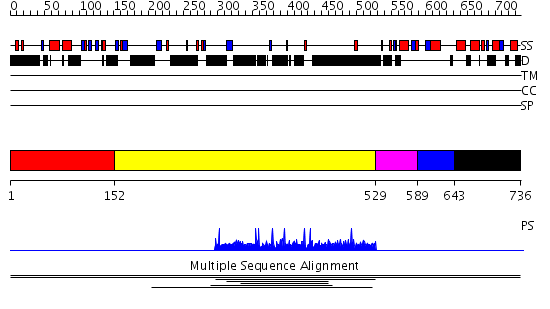
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..151] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [152..528] | 11.3 | RBP1 | |
| 3 | View Details | [529..588] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [589..642] | 9.73 | Cysteine rich protein B (HcpB) | |
| 5 | View Details | [643..736] | 9.73 | Cysteine rich protein B (HcpB) |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)