













| General Information: |
|
| Name(s) found: |
VPS27_SCHPO
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 610 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRWWNSNSQ FASDIEKATS ETLPAGSEEI SLYLEISDQI RSKSVDPKFA MRILKSRIDH 60
61 SNPNVQIMAL KLTDTCVKNG GSGFLLEIAS REFMDNLVSI LRSPAGIDED VKMVILRYIQ 120
121 SWALAVPDTN SPLSYIIHVY QNLKDGDYEF PEPSQNITSK FLDTETPPDW TDSEVCLRCR 180
181 TPFTFTNRKH HCRNCGGVFC NQCSSKTLSL PHLGINQPVR VCDSCYSLRT KPKGSKSRAR 240
241 NERKFHAKTR KTPSKPVTNN EDEDIKRAIE LSLKEMPQSR EPPSYERPSE ANVVISQDQH 300
301 LTEDEDEELK RAIAISLEEA QKSSQKDDNV TAPNNMNISY PSVPAHTVST DIRSSPFSGR 360
361 PSDNPSTLIS TADADNITLY ATLVQKLKKL PPGSIFTEYQ LQELHENMGV MRTRMMRSLG 420
421 ETMSKYNGLI QALQKLQTCM RLNDALIEQR LSSTYAHHYI DSSMDSNRSI EPEPDVISTV 480
481 RNSSTIPQAS SSSVPKIVVD SSPVTENPPS HSDVMGQKDT ISSYYSTDTD VSANVMGNRH 540
541 DEVVFSDTAS GEKNTKLNID ESTNYYNTDS IDKVGEPFDE ISSGYDDLMN GNDKQGNDIP 600
601 EVQEASLIEL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
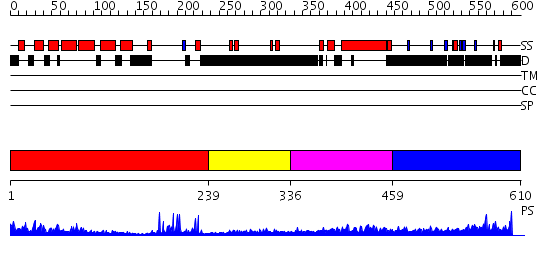
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..238] | 49.045757 | Hrs | |
| 2 | View Details | [239..335] | 3.38 | Solution Structure of Tandem UIMs of Vps27 | |
| 3 | View Details | [336..458] | 13.0 | No description for 2pjwV was found. | |
| 4 | View Details | [459..610] | 2.114996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)