













| General Information: |
|
| Name(s) found: |
pan-PH /
FBpp0099631
[FlyBase]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 747 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPHTHSRHGS SGDDLCSTDE VKIFKDEGDR EDEKISSENL LVEEKSSLID LTESEEKGHK 60
61 ISRPDHSPVF NKLDTHAPSF NMGYLVSPYS YANGSPSGLP VTMANKIGLP PFFCHNADPL 120
121 STPPPAHCGI PPYQLDPKMG LTRPALYPFA GGQYPYPMLS SDMSQVASWH TPSVYSASSF 180
181 RTPYPSSLPI NTTLASDFPF RFSPSLLPSV HATSHHVINA HSAIVGVSSK QECGVQDPTT 240
241 NNRYPRNLEA KHTSNAQSNE SKETTNDKKK PHIKKPLNAF MLYMKEMRAK VVAECTLKES 300
301 AAINQILGRR WHALGREEQA KYYELARRER QLHMQMYPDW SSRTNASRGK KRKRKQDTND 360
361 GGNNMKKCRA RFGLDQQSQW CKPCRRKKKC IRYMEALNGN GPAEDGSCFD EHGSQLSDDD 420
421 EDDYDDDKLG GSCGSADETN KIEDEDSESL NQSMPSPGCL SGLSSLQSPS TTMSLASPLN 480
481 MNANSATNVI FPASSNALLI VGADQPTAQQ RPTLVSTSGS SSGSTSSIST TPNTSSTVSP 540
541 VTCMTGPCLG SSQERAMMLG NRFSHLGMGL SPPVVSTSTS KSEPFFKPHP TVCNNPIFAL 600
601 PSIGNCSLNI SSMPNTSRNP IGANPRDINN PLSINQLTKR REYKNVELIE ASESKTIVAH 660
661 AATSIIQHVA VNGYHANHSL LNSNLGHLHH QLNNRTENPN RSEQTMLSVS NHSVNSSECH 720
721 KESDSQAIVS SNPPNAGSSD NGVISVS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
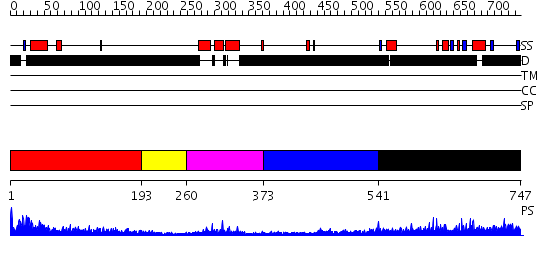
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..192] | 76.130768 | No description for PF08347.2 was found. No confident structure predictions are available. | |
| 2 | View Details | [193..259] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [260..372] | 13.522879 | Lymphoid enhancer-binding factor, LEF1 | |
| 4 | View Details | [373..540] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [541..747] | 4.218995 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)