













| General Information: |
|
| Name(s) found: |
FBpp0302678
[FlyBase]
FBpp0302677 [FlyBase] FBpp0302676 [FlyBase] CG11360-PA / FBpp0088160 [FlyBase] |
| Description(s) found:
Found 35 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 628 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC]
|
| Biological Process: |
regulation of alternative nuclear mRNA splicing, via spliceosome
[IMP]
|
| Molecular Function: |
zinc ion binding
[IEA]
protein binding [IEA] RNA binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAQVTPKIS TCINSKGSIQ KKNQAQHHDM SEQSKVNEST LPLSDDPRTL QLALELSLVG 60
61 FNDNQNCYAQ PAQPLPMPLS ARSDFEIGTI NSTLPIPSAP NCLLLPNAGA VSSEDRSKKS 120
121 QNMTECVPVP SSEHVAEIVG RQGCKIKALR AKTNTYIKTP VRGEEPVFVV TGRKEDVNKA 180
181 KREILSAADH FSLIRASRKP VSDSQNNGMV GSTTSSGGGA VPRMSGPPCM PGQVTIQVRV 240
241 PYRVVGLVVG PKGATIKHIQ QETQTYIVTP SREKEPIFEV TGLPDNVDTA RKQIEAHIAL 300
301 RTGSGSGSGD AITMQSSDSV DTKEYASLPS LTQILNDDLN SEILSSIYKN SVPSTQEYAN 360
361 NSMKTVVNVS NSVKLVQGHH TGNCFQKSEF AEMEMTANIT AADILDKRLP GNEVSVPLAK 420
421 ANVANIVFRT TSSKTLSFCP PTSTSASFHS NCNANILTRS CSSASSTTST KSTNNSANTP 480
481 PEILNIWKSI SDSIDVDEGI GDSPSIWNQP ANIIPTAHCS PTISISPTDS LLGMGEHSAN 540
541 QQNLNHAKEP IMPNLPQKIK GIQVQSNADN FLTHRECFVC NENTVTTALV PCGHNMFCME 600
601 CANHICLSMD AVCPVCNSIV YHAMRILG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
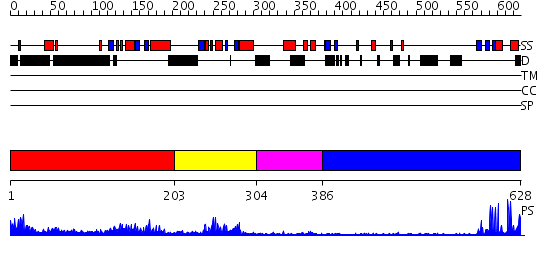
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..202] | 3.69897 | Structural genomics, 1.5A crystal structure of a hypothetical protein APE0754 from Aeropyrum pernix | |
| 2 | View Details | [203..303] | 8.30103 | No description for 2dgrA was found. | |
| 3 | View Details | [304..385] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [386..628] | 11.30103 | Cbl; N-terminal domain of cbl (N-cbl); CBL |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)